First Steps With R
This is designed to be a self-directed study session where you work through the material at your own pace. If you are at a Cookies and Code event, instructors will be on hand to help you.
What is R?
R is a free, open-source programming language that has very strong support for statistics. It was originally developed as an open source implementation of the S Programming language. It is used extensively in research and industry for areas such as data analysis, statistics, machine learning, bioinformatics, simulation, linguistics and much more.
With over 8000 freely available add-on packages that provide extensive additional functionality, R will probably have something that can help your research.
Don't just take our word for it though -- here's what others have to say
- Why use R? Five reasons - From the 'Econometrics By Simulation' blog
Installing R and RStudio
Many users of R use it from within another free piece of software called RStudio. RStudio is a powerful and productive user interface for R. It’s free and open source, and works great on Windows, Mac, and Linux.
Our first task, therefore, is to install R and RStudio.
- Install R first. Downloads are available at https://cran.rstudio.com/
- Direct link for Windows https://cran.r-project.org/bin/windows/base/
- Direct link for MacOS X https://cran.r-project.org/bin/macosx/
- Direct link for Linux https://cran.r-project.org/bin/linux/
- Install RStudio second. Downloads are available at https://www.rstudio.com/products/rstudio/download/
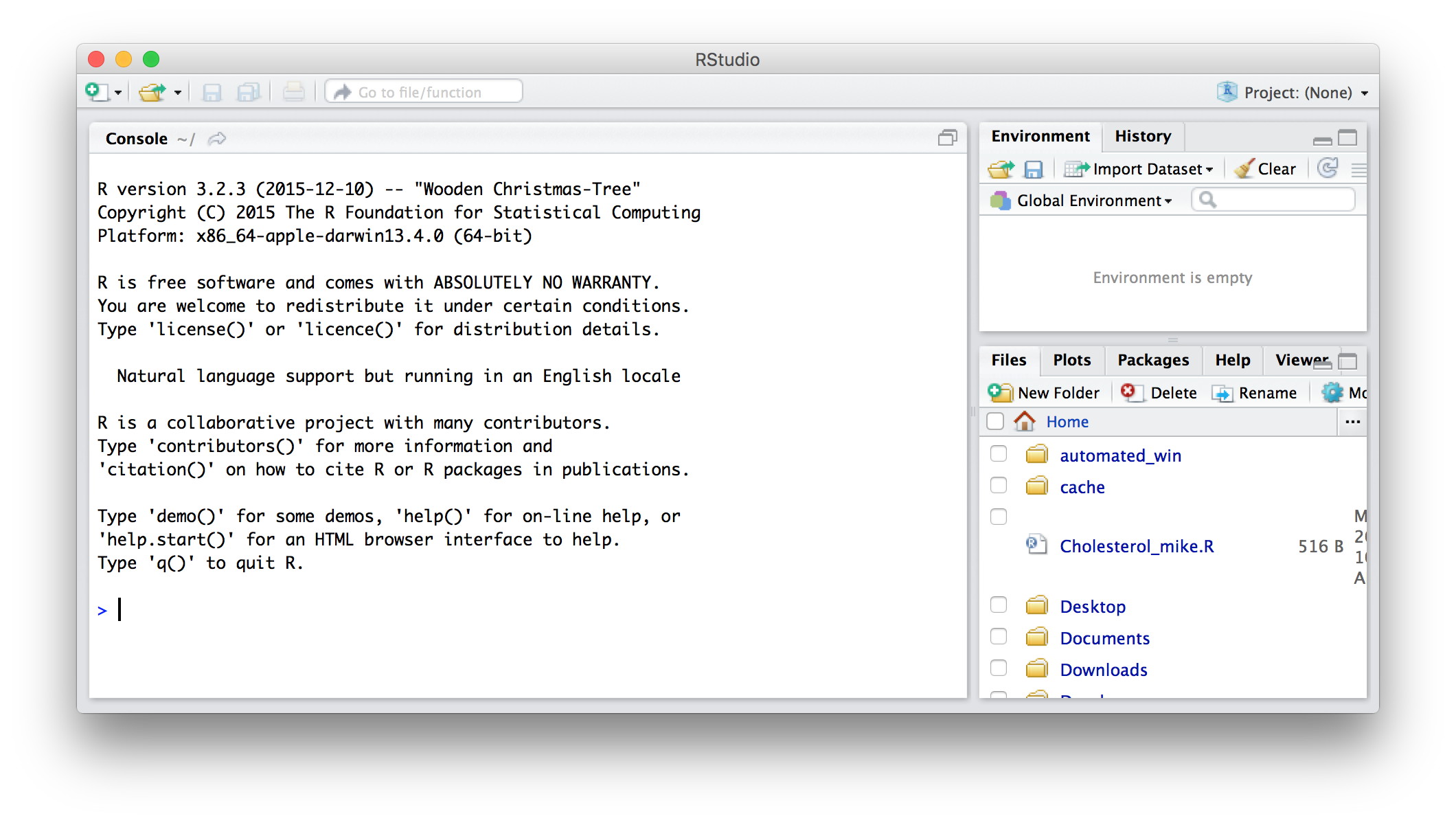
Starting RStudio
When you start RStudio, you'll be greeted with a window like the one below
R can be used interactively by typing commands into the Console panel. In this tutorial, everything that is formatted like this:
print("this is an R command")
Should be typed into the terminal. Press Return after every command.
Simple commands and calculations
R is a command based system which means that you (usually) interact with it by entering commands rather than using a Graphical User Interface (GUI). Some of these commands are rather straightforward! For example, R can be used to do arithmetic
1+1
3*9
377/120
R can also do all of the mathematical operations that you'd expect to see on a scientific calculator. For example, to take the square root of two:
sqrt(2)
This is the first time we've entered a function in R so let's discuss some details. In the above, the function name is sqrt and the function argument is 2. In R, all function arguments are enclosed in parentheses ()
R is case sensitive. For example, the correct command for square root is sqrt(2) with everything in lower case. Variations such as Sqrt(2) or SQRT(2) won't work (try it!).
R can also evaluate all the standard trigonometric functions such as sin, cos and tan. These take their arguments in radians rather than degrees. As such, a right angle is pi/2 rather than 90.
sin(pi/2)
Unlike many scientific calculators, R's log function takes the natural logarithm by default.
log(10)
If you want to calculate a logarithm to base 10, you'll need to specify the base as a second argument.
log(100,base=10)
This shows another feature of R functions -- named arguments. In this case, the named argument is base. Since the second argument to log is, by design, always the base you could have simply executed
log(100,10)
but the named argument version is more readable.
Getting help
Built in to R is a large amount of documentation that you can call on any time. For example, if you forget the details about the log function described above, ask R for help
help(log)
Variables
We'll rarely want to perform a calculation and throw away the result. It is much more likely that we'll want to store the result in R's memory for later use; either as part of future calculations or ready for export to external files.
We do this by assigning the results of calculations to variables. For example,
a <- sin(1)
b <- 10
c <- a+b
In the above, we created three variables called a, b and c. Note that as you create variables, they are shown, along with their values, in RStudio's Environment window. You can also list all variable names that currently exist in R's memory using the command
ls()
To see the value of any given variable, just type it's name followed by enter
c
To remove a variable from R's memory, we use the rm() command
rm(c)
The rm command can also remove a list of variables in one go. For example, we could remove all variables in R's memory by sending the results of ls() to it.
rm(list=ls())
Built in datasets
R comes with a package called datasets that contains a set of classic datasets such as Fisher's Iris data and Anscombe's quartet. This package is one of the few that are loaded when you start R.
To see the full list of available datasets, execute the command
library(help="datasets")
We are going to focus on the iris dataset which is stored as an R object called a Data Frame in the variable name iris. Learn more about this dataset using the help command:
help(iris)
If you run the above command, you'll see that R's documentation tells us that "iris is a data frame with 150 cases (rows) and 5 variables (columns) named Sepal.Length, Sepal.Width, Petal.Length, Petal.Width, and Species."
Let's confirm this information for ourselves by introducing a few more R commands. dim() tells us the dimensions of a data frame
dim(iris)
The names() function tells us the column names of a data frame.
names(iris)
We can extract any of the columns by name using the $ operator. To get a list of the petal lengths for example we do
iris$Petal.Length
The str() function gives a compact summary of the structure of its input
str(iris)
The head() function shows us the first 6 data points.
head(iris)
You could display the entire data frame by simply entering
iris
Alternatively, we can obtain some summary statistics about this data frame using the summary() command
summary(iris)
Plotting data
Let's extract the columns Petal.Length and Petal.Width and plot them against each other
x = iris$Petal.Length
y = iris$Petal.Width
plot(x,y)
We add axis labels and titles by supplying named arguments to the plot command
plot(x,y,xlab="Petal Length",ylab="Petal Width",main="Iris Data")
Each datapoint has an iris species associated with it - one of setosa, versicolor and virginica. We can see this by asking R what the structure of the iris$Species column is
str(iris$Species)
Factors are how R represent categorical variables. We can see what the factor levels are with
levels(iris$Species)
We can include this information on the plot by coloring each datapoint according to its species.
plot(x,y,xlab="Petal Length",ylab="Petal Width",main="Iris Data",col=iris$Species)
Finally, let's add a legend
plot(x,y,xlab="Petal Length",ylab="Petal Width",main="Iris Data",col=iris$Species)
legend(x = 1, y = 2.5, legend = levels(iris$Species), col = c(1:3), pch=1)
Exercise - Tooth growth:
Try summarising and plotting a different dataset using the commands you've learned. The name of the dataset to investigate is ToothGrowth. Again, you can use help(ToothGrowth) to see contextual information and metadata.
Packages
R has many functions built in but there are over 8000 freely available add-on packages that provide thousands more functions. Once you know the name of a package, you call install it very easily.
For example, a package called ggplot2 is widely used to create high quality graphics. To install ggplot2:
install.packages("ggplot2")
We make all of the ggplot2 functions available to our R session with the library command
library(ggplot2)
Among other things, this makes the qplot function available to us. We can use this as an alternative to the basic plot command described above
qplot(iris$Petal.Length, iris$Petal.Width,col=iris$Species)
Alternatively, we can save ourselves typing iris$ a lot by telling qplot that the data we are referring to is the iris data
qplot(data=iris,Petal.Length, Petal.Width,col=Species)
To get help about the functionality in the ggplot2 package:
help(package=ggplot2)
Exercise (Packages)
A very popular R package is MASS which was created to support the book Modern Applied Statistics with S. This contains many more classic data sets which can be used to develop your R skills.
- Install the MASS package on your machine.
- Explore the MASS package's documentation and find a dataset that interests you.
- Load the MASS library into your R session.
- Take a look at the dataset you chose in part (2) using what you've learned so far.
The current working directory
Working with built-in datasets is great for practice but for real-life work its vital that you can import our own data. Before we do this, we must learn where R is expecting to find your files. It does this using the concept of current working directory. To see what the current working directory is, execute
getwd()
You can create a new directory using dir.create()
dir.create('mydata')
Move into this new directory using setwd()
setwd('mydata')
See its contents with
dir()
The current working directory is where R is currently looking for files and also where it will put any files it creates unless you tell it otherwise.
Importing your own data
In this section, you'll learn how to import data into R from the common .csv (comma separated values) format.
Download the file example_data.csv to your current working directory. You can either do this manually, using your web browser, or you can use the R command download.file
download.file('https://raw.githubusercontent.com/mikecroucher/Code_cafe/master/First_steps_with_R/example_data.csv',destfile="example_data.csv")
Ensure that the file is in your current working directory using the dir() function
dir()
Import the .csv file using the read.csv() function
example_data <- read.csv('example_data.csv')
The variable example_data will be an R data frame -- exactly the same type of object as the iris data we looked at earlier.
Exercise - example_data
- Show the first few lines of example_data
- Create a plot of the example_data
- Show summary statistics of example_data
Scripts
In the simplest terms, a script is just a text file containing a list of R commands. We can run this list in order with a single command called source()
An alternative way to think of a script is as a permanent, repeatable, annotated, shareable, cross-platform archive1 of your analysis! Everything required to repeat your analysis is available in a single place. The only extra required ingredient is a computer.
For example, based on the article at http://www.walkingrandomly.com/?p=5254, we have created a script called best_fit.R that finds the parameters p1 and p2 such that the curve p1*cos(p2*xdata) + p2*sin(p1*xdata) is a best fit for the example_data described earlier. The details of this are beyond the scope of this course but you can easily download and run this analysis yourself.
download.file('https://raw.githubusercontent.com/mikecroucher/Code_cafe/master/First_steps_with_R/best_fit.R',destfile='best_fit.R')
source('best_fit.R')
By doing this, you have reproduced the analysis that we did. You are able to check and extend our results or apply the code to your own work. Making code and data publicly available like this is the foundation of Open Data Science
Further reading and next steps
In this session, we told you how to import data from a file but not how to export it. The following link will teach you how to export to .csv.
There are many online resources for learning R. Here are some we like
References
[1] Getting Started with R - An Introduction for Biologists. Authors: Beckerman and Petchey.