Rice D2K Lab Fall 2020 project
By: Aneel Damaraju, Chiraag Kaushik, Andrew Pham, Kunal Rai, Tucker Reinhardt, Frank Yang
Mentors: Sebastian Acosta, PhD; Mubbasheer Ahmed, MD; Parag Jain, MD
This main goal of this project is to develop an algorithm for early detection of electrical instability of the heart, specifically in pediatric patients with hypoplastic left heart syndrome. To read our report for this project check out this link!
We also created a dashboard to simulate real-time patient monitoring.
Ensure that the working directory of any python files are set to the project root
This will ensure that package imports work inside the project
Pycharm: Edit the run configuration and set the working directory to the project root
Terminal: From the project root, run export PYTHONPATH=$PYTHONPATH:'pwd' to set the working directory, then run the desired script from the project root (i.e. python3 src/<package_name>/.../<scriptname>.py
-
git clone https://github.com/RiceD2KLab/TCH_CardiacSignals_F20.git -
pip3 install -r requirements.txtinto the virtual environment
NOTE: tensorflow is not yet available for Python 3.9; please use Python 3.8.5 -
Optional (if you want to run the entire pipeline over the original data): Download the H5 files folder from the TCH box into the project root. Rename to
Data_H5_Files
We provide the notebook run.ipynb that runs a sample patient (id = 16) through our pipeline. Specifically, it runs the
patient through our preprocessing, modeling, and validation sections of the pipeline.
The notebook defaults to loading a pretrained model to avoid the expensive autoencoder training step. To get this pretrained model,
download the 3 pre-trained models from the TCH Box (in the folder called PATIENT 16 PRETRAINED MODELS) into the Working_Data directory in your cloned repo (detailed instructions inside the notebook)
For reproducibility, we add notes in the README for each module on how to reproduce our figures. Head to the srcdirectory, for information on preprocessing, modeling and validation.
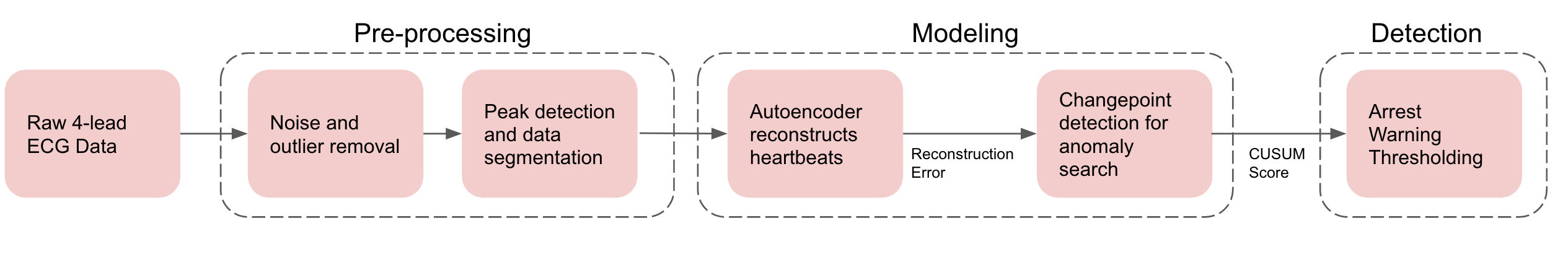
The following diagram is an overview of our data science pipeline
Data_H5_Files/- contains the raw ECG signals in the form of H5 files -> contains sample for patient 16images/- contains plots used for the final report and presentationsrc/- source code for the projectWorking_Data/- directory for intermediate data such as normalized heartbeats, reconstructions, etc.ReportPDF.pdf- Formal final report for this projectrequirements.txt- Packages required to run this repositoryrun.ipynb- notebook for running a single patient through the pipeline, including preprocessing and modeling
The data we used to develop the pipeline is in the TCH Box, located in the folder called Data H5 Files.
If you want to run the original data through the pipeline, then download the H5 files from the above folder into the Data_H5_Files folder in the repo. (Note the underscores)
If you want to run new data through the pipeline, then for each patient:
- Put 6 hours of 4-lead ECG data into an H5 file
- Name the H5 file
Reference_idx_{unique_id}_Time_block_1.h5, where{unique_id}is some unique number that is not already used within thesrc/utils/file_indexer.pyindices. - Put this file in the
Data_H5_Filesarray - Add the unique index of the file into the
new_patient_idarray
*Note: If there are any IDs in the new_patient_id array, then the pipeline defaults to reading from this array.
If the new_patient_id array is empty, then the pipeline defaults to using the original data referenced in the previous subsection.