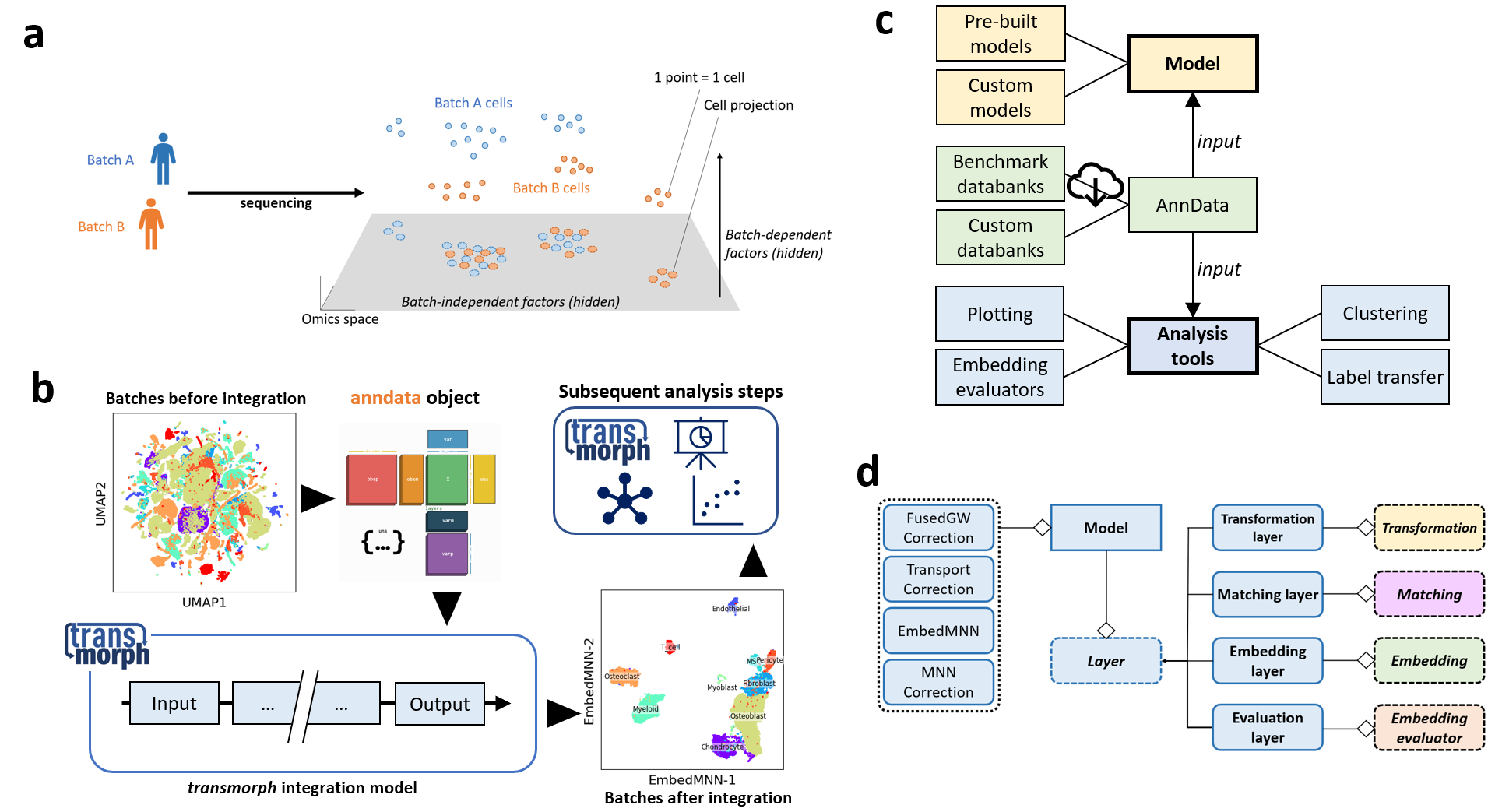
transmorph is a python framework dedicated to data integration, with a focus on single-cell applications. Dataset integration describes the problem of embedding two or more datasets together, across different batches or feature spaces, so that similar samples end up close from one another. In transmorph we aim to provide a comprehensive framework to design, apply, report and benchmark data integration models using a system of interactive building blocks supported by statistical and plotting tools. We included pre-built models as well as benchmarking databanks in order to easily set up integration tasks. This package can be used in compatibility with scanpy and anndata packages, and works in jupyter notebooks.
Transmorph is also computationally efficient, and can scale to large datasets with competitive integration quality.
Documentation
https://transmorph.readthedocs.io/en/latest/
Installation
transmorph can be installed either from source of from the python repository PyPi. PyPi version is commonly more stable, but may not contain latest features, while you can find the development version on GitHub. Using a python environment is highly recommended (for instance pipenv) in order to easily handle dependencies and versions. transmorph has only be tested for python >=3.9, on Linux and Windows systems.
See the instructions: https://transmorph.readthedocs.io/en/latest/sections/installation.html
Quick starting with a pre-built model
All transmorph models take a list or a dictionary containing AnnData objects as input for data integration. Let us start by loading some benchmarking data, gathered from [Chen 2020] (3.4GB size).
from transmorph.datasets import load_chen_10x
chen_10x = load_chen_10x()One can then either create a custom integration model, or load a pre-built transmorph model. We will choose the EmbedMNN model with default parameters for this example, which embeds all datasets into a common abstract 2D space.
from transmorph.models import EmbedMNN
model = EmbedMNN()
model.transform(chen_10x)Integration embedding coordinates can be gathered in each AnnData object, in AnnData.obsm['X_transmorph'].
chen_10x['P01'].obsm['X_transmorph']One can for instance use a plotting function from transmorph to display integration results.
from transmorph.utils.plotting import scatter_plot
scatter_plot(chen_10x, use_rep="X_transmorph")
scatter_plot(chen_10x, use_rep="X_transmorph", color_by="class")Citing transmorph
If you find transmorph useful for your research, please consider citing our pre-print which can be found on bioRxiv.
@article{fouche2022transmorph,
title={transmorph: a unifying computational framework for single-cell data integration},
author={Fouch{\'e}, Aziz, Chadoutaud, Lo{\¨i}c, Delattre, Olivier and Zinovyev, Andrei},
journal={bioRxiv},
year={2022}
}