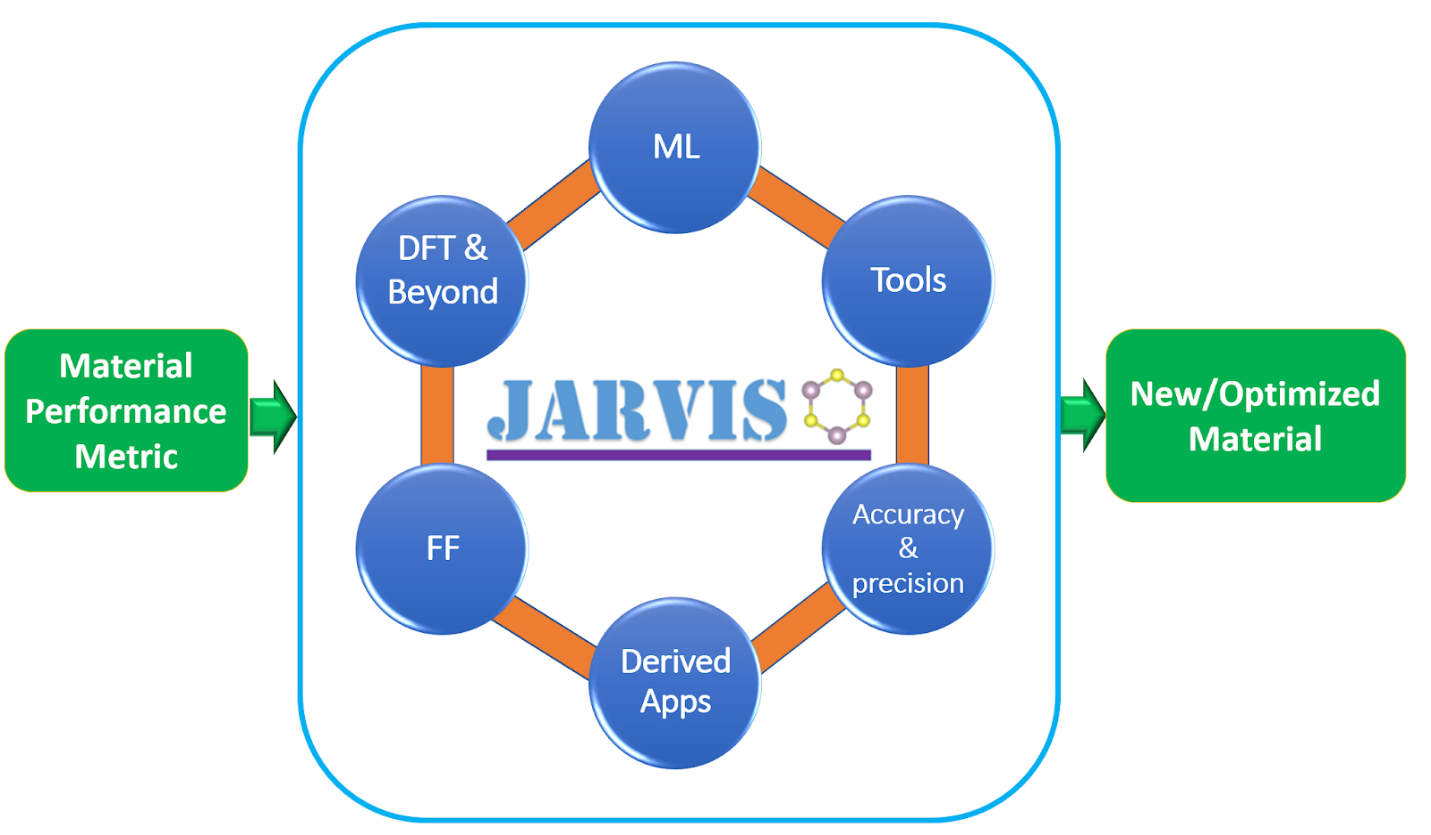
The JARVIS-Tools is an open-access software package for atomistic data-driven materials desgin. JARVIS-Tools can be used for a) setting up calculations, b) analysis and informatics, c) plotting, d) database development and e) web-page development.
JARVIS-Tools empowers NIST-JARVIS (Joint Automated Repository for Various Integrated Simulations) repository which is an integrated framework for computational science using density functional theory, classical force-field/molecular dynamics and machine-learning. The NIST-JARVIS official website is: https://jarvis.nist.gov . This project is a part of the Materials Genome Initiative (MGI) at NIST (https://mgi.nist.gov/).
For more details, checkout our latest article: The joint automated repository for various integrated simulations (JARVIS) for data-driven materials design and YouTube videos
- Software workflow tasks for preprcessing, executing and post-processing: VASP, Quantum Espresso, Wien2k BoltzTrap, Wannier90, LAMMPS, Scikit-learn, TensorFlow, LightGBM, Qiskit, Tequila, Pennylane, DGL, PyTorch.
- Several examples: Notebooks and test scripts to explain the package.
- Several analysis tools: Atomic structure, Electronic structure, Spacegroup, Diffraction, 2D materials and other vdW bonded systems, Mechanical, Optoelectronic, Topological, Solar-cell, Thermoelectric, Piezoelectric, Dielectric, STM, Phonon, Dark matter, Wannier tight binding models, Point defects, Heterostructures, Magnetic ordering, Images, Spectrum etc.
- Database upload and download: Download JARVIS databases such as JARVIS-DFT, FF, ML, WannierTB, Solar, STM and also external databases such as Materials project, OQMD, AFLOW etc.
- Access raw input/output files: Download input/ouput files for JARVIS-databases to enhance reproducibility.
- Train machine learning models: Use different descriptors, graphs and datasets for training machine learning models.
- HPC clusters: Torque/PBS and SLURM.
- Available datasets: Summary of several datasets .
>>> pip install -U jarvis-tools
or
>>> conda install -c conda-forge jarvis-tools
For detailed instructions, please see Installation instructions
>>> from jarvis.core.atoms import Atoms >>> box = [[2.715, 2.715, 0], [0, 2.715, 2.715], [2.715, 0, 2.715]] >>> coords = [[0, 0, 0], [0.25, 0.25, 0.25]] >>> elements = ["Si", "Si"] >>> Si = Atoms(lattice_mat=box, coords=coords, elements=elements) >>> density = round(Si.density,2) >>> print (density) 2.33 >>> >>> from jarvis.db.figshare import data >>> dft_3d = data(dataset='dft_3d') >>> print (len(dft_3d)) 48527 >>> from jarvis.io.vasp.inputs import Poscar >>> for i in dft_3d: ... atoms = Atoms.from_dict(i['atoms']) ... poscar = Poscar(atoms) ... jid = i['jid'] ... filename = 'POSCAR-'+jid+'.vasp' ... poscar.write_file(filename) >>> dft_2d = data(dataset='dft_2d') >>> print (len(dft_2d)) 1070 >>> for i in dft_2d: ... atoms = Atoms.from_dict(i['atoms']) ... poscar = Poscar(atoms) ... jid = i['jid'] ... filename = 'POSCAR-'+jid+'.vasp' ... poscar.write_file(filename) >>> # Example to parse DOS data from JARVIS-DFT webpages >>> from jarvis.db.webpages import Webpage >>> from jarvis.core.spectrum import Spectrum >>> import numpy as np >>> new_dist=np.arange(-5, 10, 0.05) >>> all_atoms = [] >>> all_dos_up = [] >>> all_jids = [] >>> for ii,i in enumerate(dft_3d): all_jids.append(i['jid']) ... try: ... w = Webpage(jid=i['jid']) ... edos_data = w.get_dft_electron_dos() ... ens = np.array(edos_data['edos_energies'].strip("'").split(','),dtype='float') ... tot_dos_up = np.array(edos_data['total_edos_up'].strip("'").split(','),dtype='float') ... s = Spectrum(x=ens,y=tot_dos_up) ... interp = s.get_interpolated_values(new_dist=new_dist) ... atoms=Atoms.from_dict(i['atoms']) ... ase_atoms=atoms.ase_converter() ... all_dos_up.append(interp) ... all_atoms.append(atoms) ... all_jids.append(i['jid']) ... filename=i['jid']+'.cif' ... atoms.write_cif(filename) ... break ... except Exception as exp : ... print (exp,i['jid']) ... pass
Find more examples at
Please cite the following if you happen to use JARVIS-Tools for a publication.
https://www.nature.com/articles/s41524-020-00440-1
Choudhary, K. et al. The joint automated repository for various integrated simulations (JARVIS) for data-driven materials design. npj Computational Materials, 6(1), 1-13 (2020).
Please see Publications related to JARVIS-Tools
Please report bugs as Github issues (https://github.com/usnistgov/jarvis/issues) or email to kamal.choudhary@nist.gov.
NIST-MGI (https://www.nist.gov/mgi).
Please see Code of conduct
jarvis/
├── ai
│ ├── descriptors
│ │ ├── cfid.py
│ │ ├── coulomb.py
│ ├── gcn
│ ├── pkgs
│ │ ├── lgbm
│ │ │ ├── classification.py
│ │ │ └── regression.py
│ │ ├── sklearn
│ │ │ ├── classification.py
│ │ │ ├── hyper_params.py
│ │ │ └── regression.py
│ │ └── utils.py
│ ├── uncertainty
│ │ └── lgbm_quantile_uncertainty.py
├── analysis
│ ├── darkmatter
│ │ └── metrics.py
│ ├── defects
│ │ ├── surface.py
│ │ └── vacancy.py
│ ├── diffraction
│ │ └── xrd.py
│ ├── elastic
│ │ └── tensor.py
│ ├── interface
│ │ └── zur.py
│ ├── magnetism
│ │ └── magmom_setup.py
│ ├── periodic
│ │ └── ptable.py
│ ├── phonon
│ │ ├── force_constants.py
│ │ └── ir.py
│ ├── solarefficiency
│ │ └── solar.py
│ ├── stm
│ │ └── tersoff_hamann.py
│ ├── structure
│ │ ├── neighbors.py
│ │ ├── spacegroup.py
│ ├── thermodynamics
│ │ ├── energetics.py
│ ├── topological
│ │ └── spillage.py
├── core
│ ├── atoms.py
│ ├── composition.py
│ ├── graphs.py
│ ├── image.py
│ ├── kpoints.py
│ ├── lattice.py
│ ├── pdb_atoms.py
│ ├── specie.py
│ ├── spectrum.py
│ └── utils.py
├── db
│ ├── figshare.py
│ ├── jsonutils.py
│ ├── lammps_to_xml.py
│ ├── restapi.py
│ ├── vasp_to_xml.py
│ └── webpages.py
├── examples
│ ├── lammps
│ │ ├── jff_test.py
│ │ ├── Al03.eam.alloy_nist.tgz
│ ├── vasp
│ │ ├── dft_test.py
│ │ ├── SiOptb88.tgz
├── io
│ ├── boltztrap
│ │ ├── inputs.py
│ │ └── outputs.py
│ ├── calphad
│ │ └── write_decorated_poscar.py
│ ├── lammps
│ │ ├── inputs.py
│ │ └── outputs.py
│ ├── pennylane
│ │ ├── inputs.py
│ ├── phonopy
│ │ ├── fcmat2hr.py
│ │ ├── inputs.py
│ │ └── outputs.py
│ ├── qe
│ │ ├── inputs.py
│ │ └── outputs.py
│ ├── qiskit
│ │ ├── inputs.py
│ ├── tequile
│ │ ├── inputs.py
│ ├── vasp
│ │ ├── inputs.py
│ │ └── outputs.py
│ ├── wannier
│ │ ├── inputs.py
│ │ └── outputs.py
│ ├── wanniertools
│ │ ├── inputs.py
│ │ └── outputs.py
│ ├── wien2k
│ │ ├── inputs.py
│ │ ├── outputs.py
├── tasks
│ ├── boltztrap
│ │ └── run.py
│ ├── lammps
│ │ ├── templates
│ │ └── lammps.py
│ ├── phonopy
│ │ └── run.py
│ ├── vasp
│ │ └── vasp.py
│ ├── queue_jobs.py
├── tests
│ ├── testfiles
│ │ ├── ai
│ │ ├── analysis
│ │ │ ├── darkmatter
│ │ │ ├── defects
│ │ │ ├── elastic
│ │ │ ├── interface
│ │ │ ├── magnetism
│ │ │ ├── periodic
│ │ │ ├── phonon
│ │ │ ├── solar
│ │ │ ├── stm
│ │ │ ├── structure
│ │ │ ├── thermodynamics
│ │ │ ├── topological
│ │ ├── core
│ │ ├── db
│ │ ├── io
│ │ │ ├── boltztrap
│ │ │ ├── calphad
│ │ │ ├── lammps
│ │ │ ├── pennylane
│ │ │ ├── phonopy
│ │ │ ├── qiskit
│ │ │ ├── qe
│ │ │ ├── tequila
│ │ │ ├── vasp
│ │ │ ├── wannier
│ │ │ ├── wanniertools
│ │ │ ├── wien2k
│ │ ├── tasks
│ │ │ ├── test_lammps.py
│ │ │ └── test_vasp.py
└── README.rst