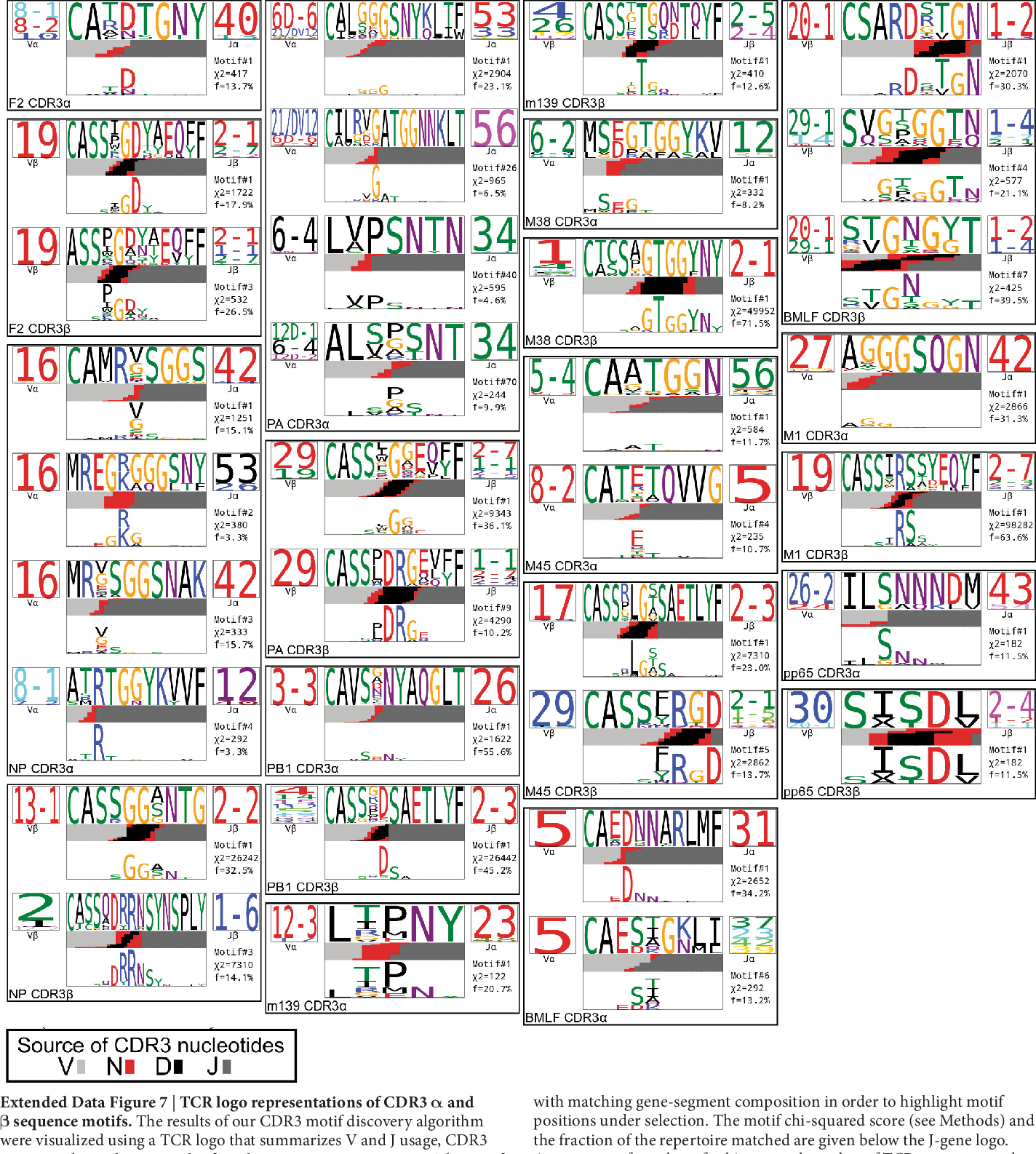
Suppose you've done some flashywork with tcrdist3, our research group's open-source Python package that enables a broad array of flexible T cell receptor sequence analyses; however, you are old-school and you want regex motif patterns and the classic motif plots as they were developed in the original TCRdist 2017 scripts suite associated with the Dash et al. Nature (2017) doi:10.1038/nature22383 manuscript.
pip install git+https://github.com/kmayerb/tcrregex.git
After install, download key background files. THIS IS NOT OPTIONAL.
python -c "import tcrregex as td; td.install_test_files.install_test_files()"
python -c "import tcrregex as td; td.setup_db.install_all_next_gen()"
docker pull quay.io/kmayerb/tcrregex:0.1.0
If using the Docker image, the next-gen files (see above section on Data File) are already available in the Docker image; however, you will need to pip install git+https://github.com/kmayerb/tcrdist3.git@0.1.4 if want to follow the usage example.
First, with tcrdist3:
import pandas as pd
from tcrdist.repertoire import TCRrep
df = pd.read_csv("dash.csv")
tr = TCRrep(cell_df = df,
organism = 'mouse',
chains = ['alpha','beta'],
db_file = 'alphabeta_gammadelta_db.tsv')Now, boot up the tcrregex package
from tcrregex.mappers import populate_legacy_fields
from tcrregex.subset import TCRsubset
from tcrregex.cdr3_motif import TCRMotif
from tcrregex.storage import StoreIOMotif, StoreIOEntropy
from tcrregex import plotting
# define a logical criteria
ind = (tr.clone_df.epitope == "PA")
# subset the TCRrep clone DataFrame to only those sequences meeting that criteria
clone_df_subset = tr.clone_df[ind].reset_index(drop = True).copy()
# subset the alpha chain and beta chain distance matrices using the `clone_df_subset.clone_id` index
dist_a_subset = pd.DataFrame(tr.pw_alpha[ind,:][:,ind])
dist_b_subset = pd.DataFrame(tr.pw_beta[ind,:][:,ind])
# Check dimensions and shapes of inputs
assert dist_a_subset.shape[0] == clone_df_subset.shape[0]
assert dist_a_subset.shape[1] == clone_df_subset.shape[0]
assert dist_b_subset.shape[0] == clone_df_subset.shape[0]
assert dist_b_subset.shape[1] == clone_df_subset.shape[0]
assert isinstance(dist_a_subset, pd.DataFrame)
assert isinstance(dist_b_subset, pd.DataFrame)
assert isinstance(clone_df_subset, pd.DataFrame)
# Use the populate_legacy_fields function to add some columns needed for compatability with tcrdist1
clone_df_subset = populate_legacy_fields(df = clone_df_subset, chains =['alpha', 'beta'])
# FOR DEMO ONLY: Limit the sarch to first 50 seqs
clone_df_subset = clone_df_subset.iloc[0:50, :].copy()
dist_b_subset = dist_b_subset.iloc[0:50, 0:50].copy()
dist_a_subset = dist_a_subset.iloc[0:50, 0:50].copy()
# initialize an instance of the TCRsubset class.
ts = TCRsubset(clone_df_subset,
organism = "mouse",
epitopes = ["PA"] ,
epitope = "PA",
chains = ["A","B"],
dist_a = dist_a_subset,
dist_b = dist_b_subset)
# Chilax this step can take forever!
ts.find_motif()
# So make sure to save your motifs DataFrame
ts.motif_df.to_csv("saved_motifs.csv", index = False)
# You can always reload these and skip the wait
ts.motif_df = pd.read_csv("saved_motifs.csv")
# Output alpha-chain motifs
motif_list_a = list()
motif_logos_a = list()
for i,row in ts.motif_df[ts.motif_df.ab == "A"].iterrows():
StoreIOMotif_instance = ts.eval_motif(row)
motif_list_a.append(StoreIOMotif_instance)
motif_logos_a.append(plotting.plot_pwm(StoreIOMotif_instance, create_file = False, my_height = 200, my_width = 600))
with open('alpha_motifs.html' , 'w') as outfile:
outfile.write('<html>\n')
for motif in motif_list_a:
svg = plotting.plot_pwm(motif, create_file = False, my_height = 200, my_width = 600)
outfile.write(f"<div>{svg}</div>")
outfile.write('</html>\n')
# Output beta-chain motifs
motif_list_b = list()
motif_logos_b = list()
for i,row in ts.motif_df[ts.motif_df.ab == "B"].iterrows():
StoreIOMotif_instance = ts.eval_motif(row)
motif_list_b.append(StoreIOMotif_instance)
motif_logos_b.append(plotting.plot_pwm(StoreIOMotif_instance, create_file = False, my_height = 200, my_width = 600))
with open('beta_motifs.html' , 'w') as outfile:
outfile.write('<html>\n')
for motif in motif_list_b:
svg = plotting.plot_pwm(motif, create_file = False, my_height = 200, my_width = 600)
outfile.write(f"<div>{svg}</div>")
outfile.write('</html>\n')Example Output Files: