Tapping culture collections for fungal endophytes: first genome assemblies for three genera and five species in the Ascomycota
Bioinformatics analysis pipeline for:
Hill et al. (2023) Tapping culture collections for fungal endophytes: first genome assemblies for three genera and five species in the Ascomycota. Genome Biology and Evolution 15(3):evad038. doi:10.1093/gbe/evad038
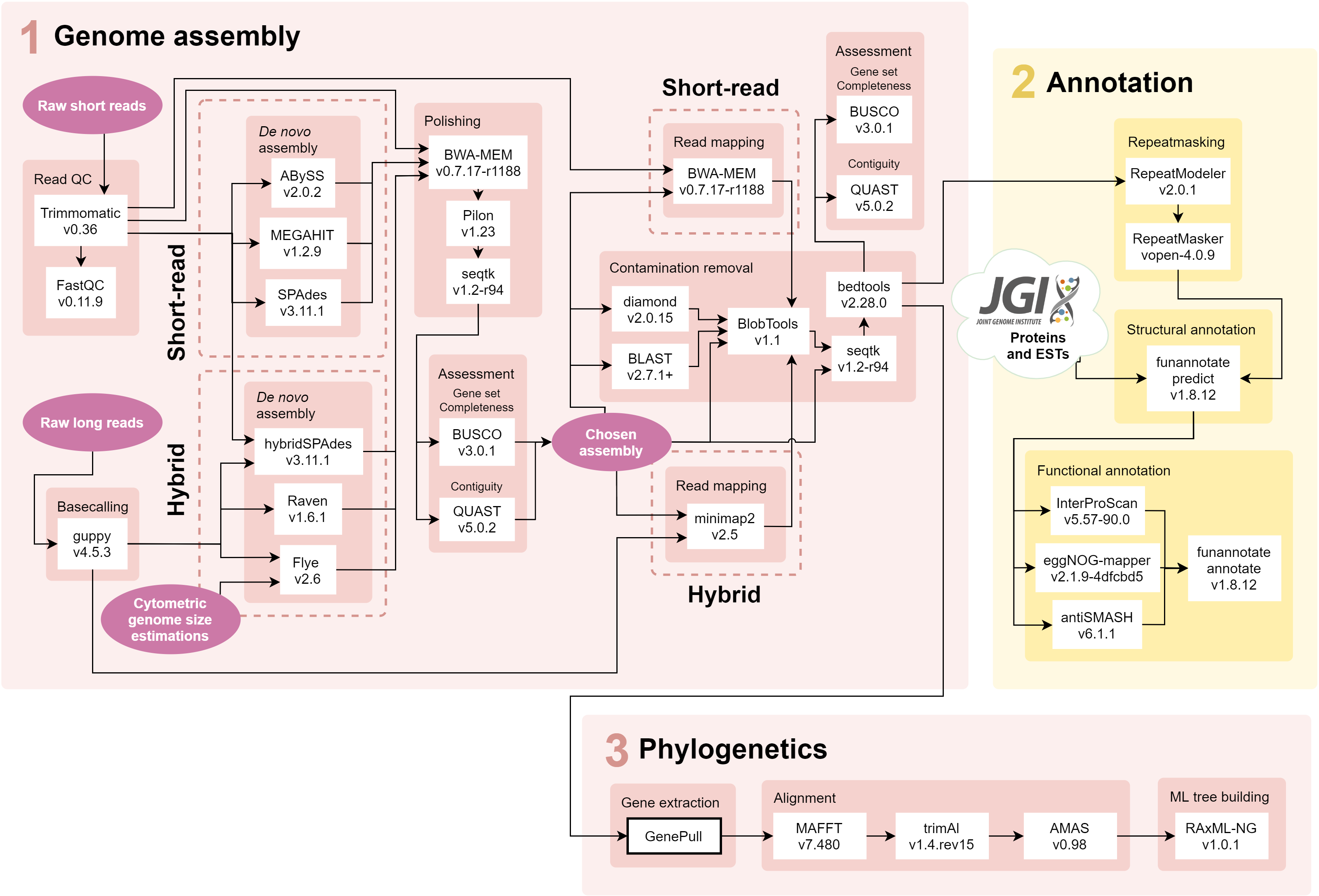
The pipeline was written for and run on Queen Mary University of London's Apocrita HPC facility which uses the Univa Grid Engine batch-queue system. This means that many of the bash scripts (.sh file endings) specify core allocation, run times and memory usage allocation that may need to be adapted for different platforms.
cd reads
qsub trimmomatic.shtrims raw reads using Trimmomatic; requiresNexteraPE-PE.fafile with adapter sequences downloaded from here (for Illumina NovaSeq 6000 151bp paired-end reads).qsub fastqc.shchecks trimmed read quality with FastQC.
qsub guppy.sh performs fast basecalling of raw MinION read data.
cd denovo_assembly
./submit_assembly.shmakes new directory and submits job scripts for each assembly tool - short-read toolsabyss.sh(ABySS),megahit.sh(MEGAHIT) andspades.sh(SPAdes), and long-read toolsflye.sh(Flye),raven.sh(Raven) andspades_hybrid(hybridSPAdes). For all tools except for ABySS, these scripts also include short-read mapping with BWA-MEM for polishing with Pilon and remove sequences <200bp using Seqtk for NCBI compliance.qsub -t 1-8 abyss_comp.shcompares the assembly stats to choose 'best' kmer size for ABySS (must be done afterabyss.shhas finished for all kmer sizes and strains), followed by short-read polishing with Pilon.
cd assessment
./submit_assessmentsubmits scripts for assembly quality statistics -quast.sh(QUAST) andbusco.sh(BUSCO), which requires the ascomycota_odb10.2020-09-10 BUSCO dataset downloaded from here) - andblast.sh(BLAST),diamond.sh(DIAMOND) andread_mapping.sh, which maps reads with BWA-MEM, to produce input for BlobTools.qsub -t 1-15 blobtools.shsubmitsblobtools.shto run BlobTools (must be done afterblast.sh,diamond.shandread_mapping.shhave finished for the strain(s) in question).
qsub -t 1-15 remove_contam.shremoves contigs which BlobTools flagged as belonging to the wrong taxonomic class using Seqtk.qsub -t 1-15 ncbi_filter.shremoves or trims contigs flagged as mitochondrial or adapter contaminations during NCBI submission with the help of bedtools; requires strain_ncbi_remove.txt and strain_ncbi_trim.bed files.
qsub quast_final.shreruns QUAST on the contaminant-filtered assemblies.mkdir busco_final | qsub -t 1-15 busco_final.shmakes a new directory and reruns BUSCO on the contaminant-filtered assemblies.qsub -t 1-15 read_mapping_final.shperforms a final round of read mapping and produces mapping statisics with SAMtools to calculate both short- and long-read coverage.
cd annotation
cd annotation/repeat_masking
qsub -t 1-15 repeatmodelermakes custom repeat library for each strain using RepeatModeler.qsub -t 1-15 repeatmasker.shuses the custom repeat libraries to softmask assemblies using RepeatMasker.
cd annotation/structural
qsub -t 1-15 funannotate.sh sorts and relabels contigs in the repeatmasked assembly before predicting gene models using funannotate. Requires protein and EST evidence downloaded from Mycocosm to be saved in this folder.
cd annotation/functional
qsub -t 1-15 eggnogmapper.shsubmits eggNOG-mapper on predicted gene models.qsub -t 1-15 antismash.shsubmits antiSMASH on predicted gene models.qsub -t 1-15 interproscan.shsubmits InterProScan on predicted gene models.qsub -t 1-15 funannotate_annotate.shmaps results from the previous three programmes onto the structural annotation and produces the.sqnfiles for NCBI submission.
cd phylogenetics
This folder contains a file - lineages - listing the 10 lineages for which trees must be built and the strains that belong to said lineages, and a file - markers - listing the 13 genetic markers selected for building the trees.
cd GenePull
./genepull.sh submits GenePull to extract selected genetic markers from contaminant filtered assemblies of each strain. Requires fasta files containing a single example sequence from a closely related taxon for each genetic marker being extracted.
file_prep.shcontains example one-liners for formatting sequence headers in each of the gene alignment fasta files so that they are identical across different genes (i.e. removing GenBank accessions; removing misc text after taxon names/vouchers; replacing spaces with underscores etc).qsub -t 1-10 align.shsubmits gene alignments using MAFFT and trimming using trimAl for each of the 10 lineages.- Gene alignments are manually checked with AliView.
qsub -t 1-10 concat.shsubmits concatenation of all gene alignments with AMAS for each of the 10 lineages.
qsub -t 1-10 raxmlng.sh submits RAxML-NG with bootstrapping until convergence or up to 1,000 replicates (whichever first) for each of the 10 lineages.
plots.r
Hill et al. (2023) Tapping culture collections for fungal endophytes: first genome assemblies for three genera and five species in the Ascomycota. Genome Biology and Evolution 15(3):evad038. doi:10.1093/gbe/evad038