LSSANet
This repository includes codes, models, and test results for our paper: "LSSANet: A Long Short Slice-Aware Network for Pulmonary Nodule Detection", MICCAI 2022 (Early Accept). This project is licensed for non-commerical research purpose only.
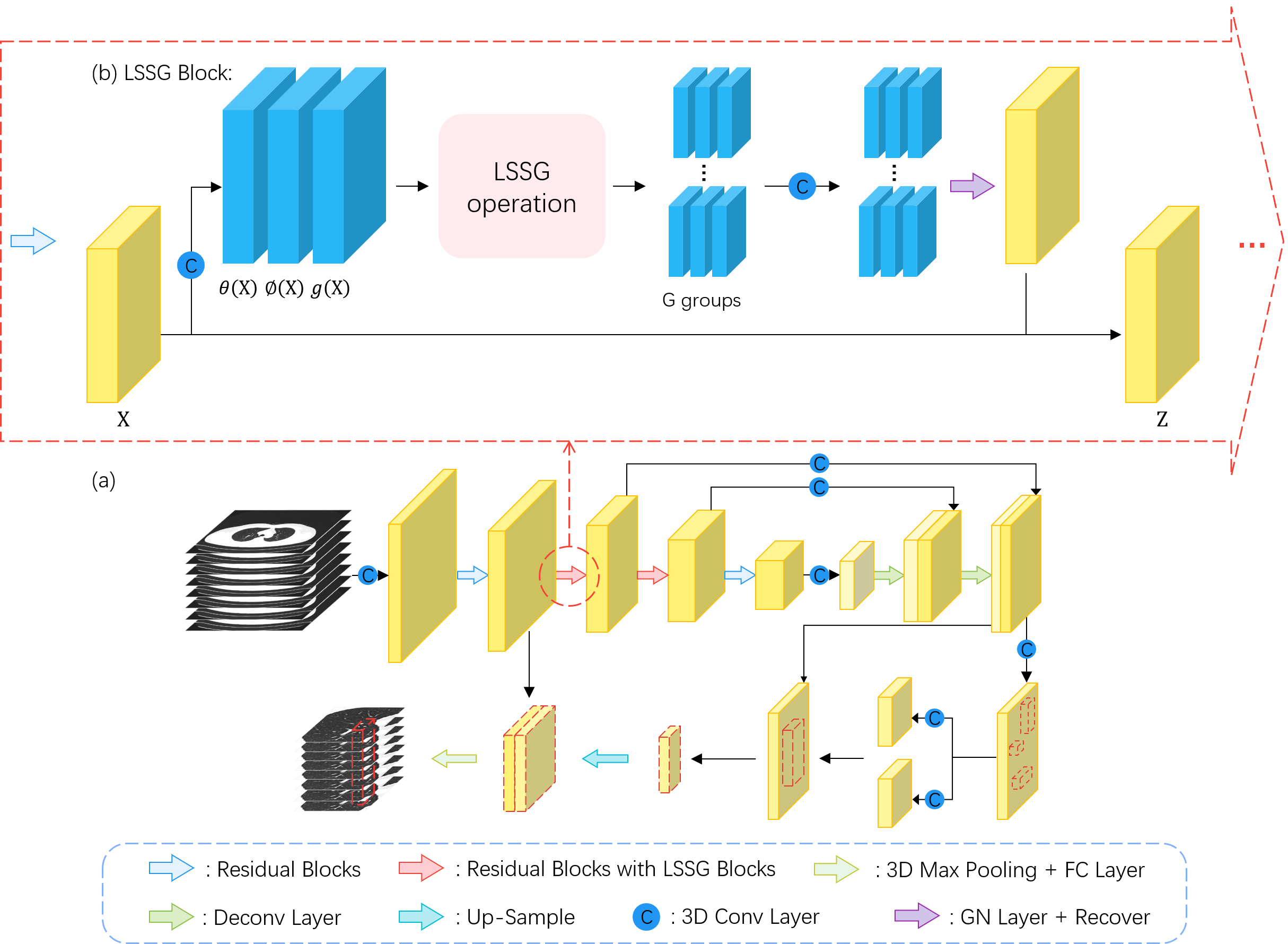
Method
Results and Models
| Method | 0.125 | 0.25 | 0.5 | 1.0 | 2.0 | 4.0 | 8.0 | Avg | Pre-trained Model |
|---|---|---|---|---|---|---|---|---|---|
| LSSANet | 51.59 | 51.59 | 58.18 | 66.88 | 77.33 | 85.35 | 89.87 | 68.69 | model & res |
Requirements
The code is built with the following libraries:
Besides, you need to install a custom module for bounding box NMS and overlap calculation.
cd build/box
python setup.py install
Data
Please refer to PN9 for data preparation and then add the data information to
single_config.py.
Testing
Run the following scripts to evaluate the model and obtain the results of FROC analysis.
python single_test.py --ckpt='./results/model/model.ckpt' --out_dir='./results/'
Training
This implementation supports multi-gpu, data_parallel training.
Change training configuration and data configuration in single_config.py, especially the path to preprocessed data.
Run the training script:
python single_train.py
Citations
If you are using the code/model/data provided here in a publication, please consider citing:
@inproceedings{DBLP:conf/miccai/XuLDKY22,
author = {Rui Xu and Yong Luo and Bo Du and Kaiming Kuang and Jiancheng Yang},
editor = {Linwei Wang and Qi Dou and P. Thomas Fletcher and Stefanie Speidel and Shuo Li},
title = {LSSANet: {A} Long Short Slice-Aware Network for Pulmonary Nodule Detection},
booktitle = {MICCAI 2022},
series = {LNCS},
volume = {13431},
pages = {664--674},
publisher = {Springer, Cham},
year = {2022},
doi = {10.1007/978-3-031-16431-6\_63}
}
Contact
For any questions, please contact: rui.xu AT whu.edu.cn.