This is the repo for paper "Improving Melanoma Detection Through Clinical Knowledge-Based Topological Graphs and Data-Driven Quantification Standards"
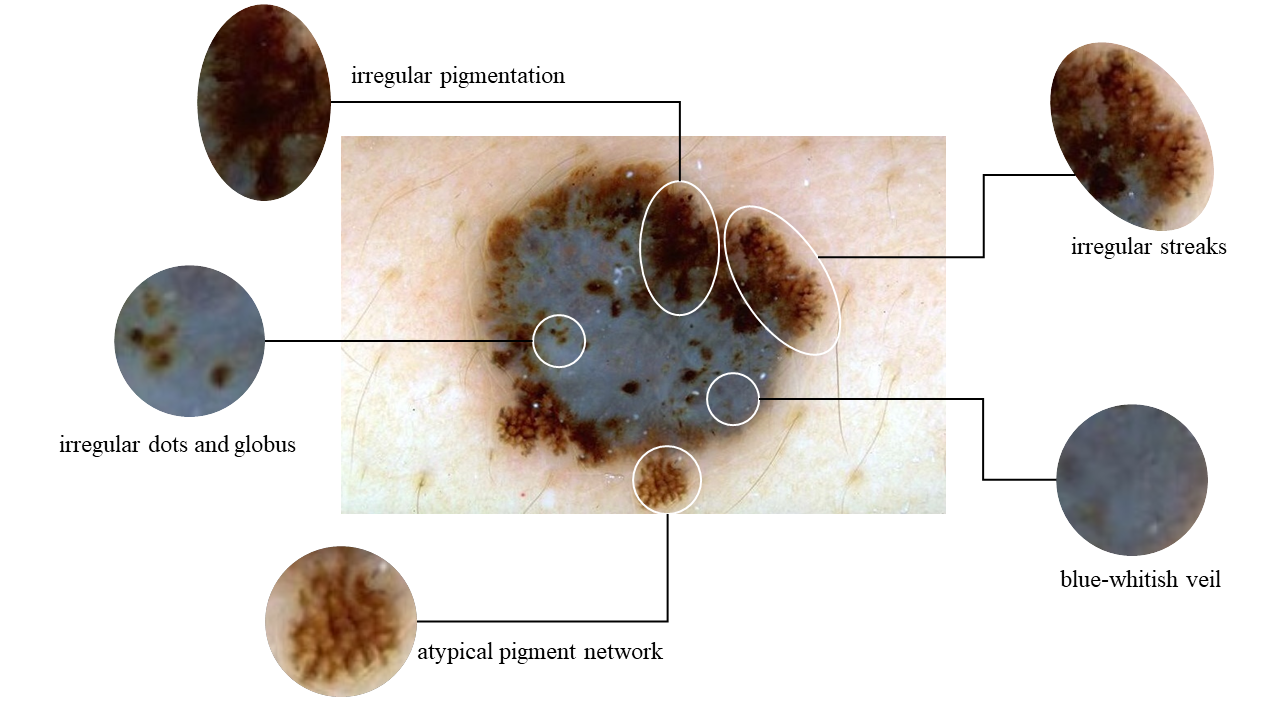
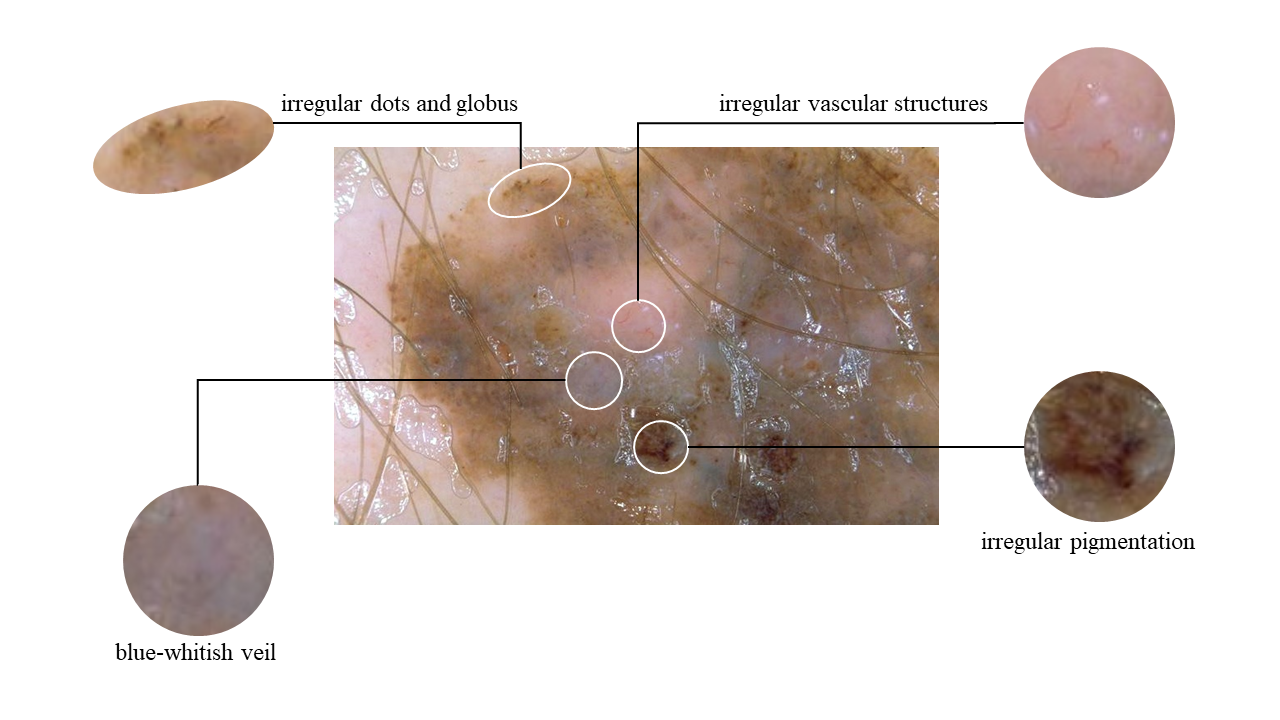
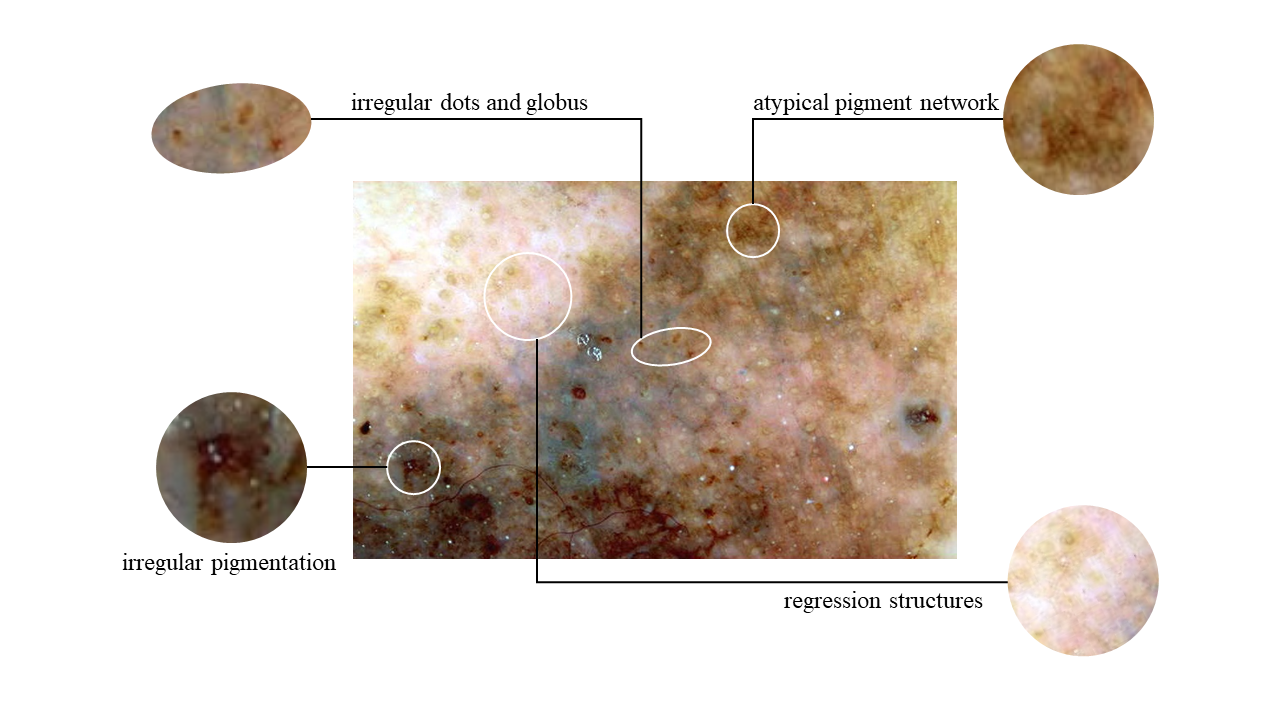
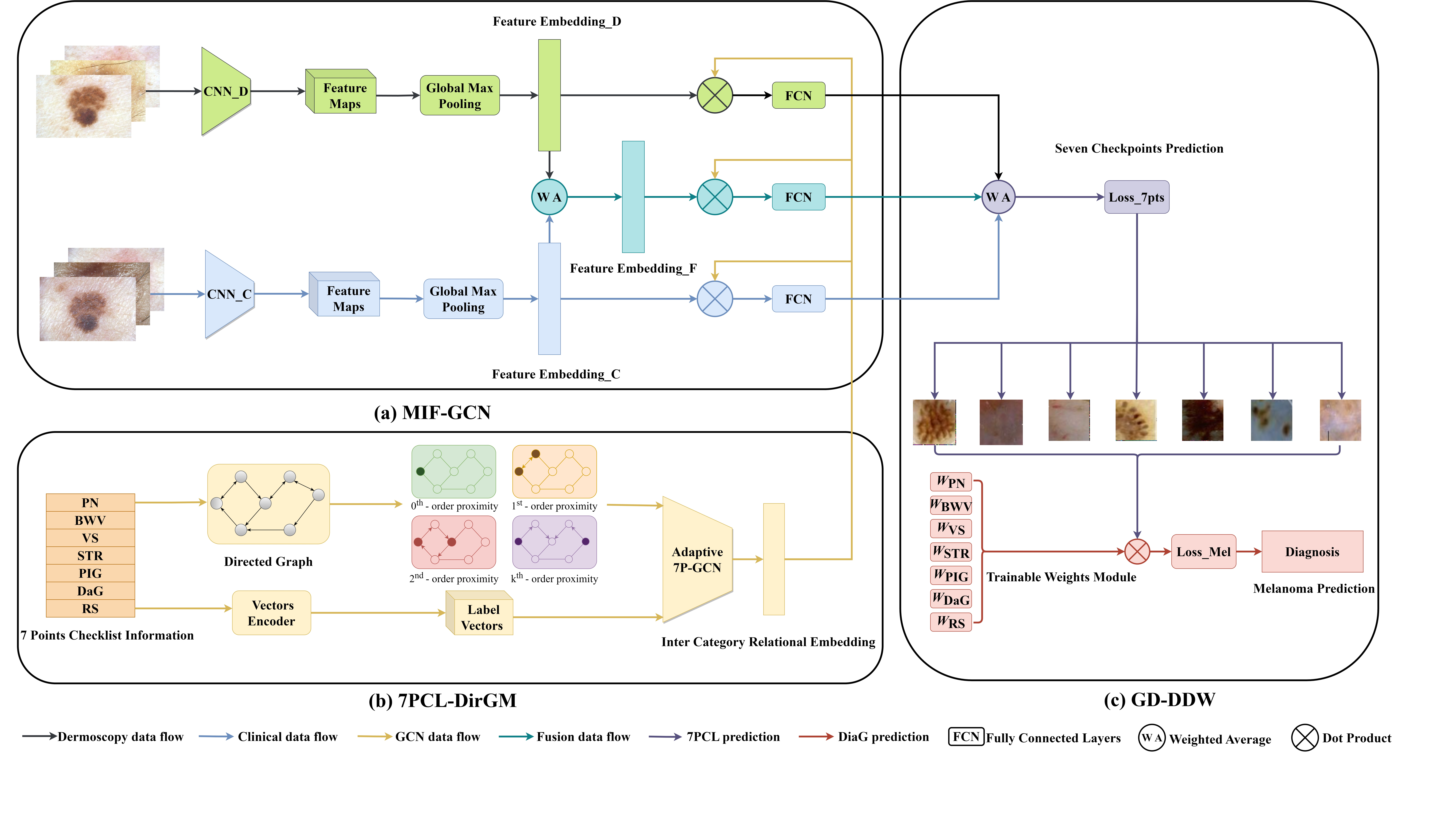
The widely utilized 7-point checklist for dermoscopy is essential for identifying malignant melanoma lesions requiring immediate attention. It assigns specific point values to seven characteristics, with major attributes worth two points and minor ones one point. A total score of three or higher prompts further evaluation, often involving a biopsy. However, a notable limitation of the current algorithm is its uniform weighting of attributes, leading to imprecision and neglecting complex interconnections. In addition, previous research predominantly focused on attribute co-occurrence rates and ordering constraints, hindering a comprehensive understanding of pathological causality. To address these limitations, we introduce a novel diagnostic approach that harmoniously blends two innovative elements: a Clinical Knowledge-Based Topological Graph (CKTG) and a Gradient Diagnostic Strategy with Data-Driven Weighting Standards (GD-DDW). The CKTG seamlessly integrates 7PCL attributes with diagnostic information, revealing both internal and external associations. By employing adaptive receptive domains and weighted edges, we establish critical connections among melanoma’s relevant features. Simultaneously, GDDDW mirrors dermatologists’ diagnostic behavior to predict melanoma progressively, significantly enhancing predictability and interoperability. Additionally, our model employs two imaging modalities for the same lesion, ensuring comprehensive feature acquisition. Our proposed algorithm shows outstanding performance in predicting malignant melanoma and its features, achieving an average AUC value of 84.8%. This was validated on the EDRA dataset, the largest publicly available dataset for the 7-point checklist algorithm. Specifically, the integrated weighting system holds the potential to provide clinicians with valuable datadriven benchmarks for their evaluations.
- compile the docker file
cd docker
docker build -t 7pgd .
- config the script and create docker container
docker create --name <container_name> -it
--shm-size=32G
-p 6006:6006
-p 8080:8080
-e DISPLAY=unix$DISPLAY
-e GDK_SCALE
-e GDK_DPI_SCALE
-v /dev/video0:/dev/video0
-v /dev/video1:/dev/video1
-v /tmp/.X11-unix:/tmp/.X11-unix
-v <proj_path>:/root/code
-v /mnt/f/datasets:/root/data
--gpus all
<docker_image>
/bin/zsh
This repository is licensed under the MIT License. See the LICENSE file for details.
Acknowledge any funding sources, contributors, or other relevant parties.
For any questions or issues, please contact First Author or open an issue in this repository.