CBioNAMER (Nested nAMed Entity Recognition for Chinese Biomedical Text) is our method used in CBLUE1.0 (Chinese Biomedical Language Understanding Evaluation), a benchmark of Nested Named Entity Recognition.
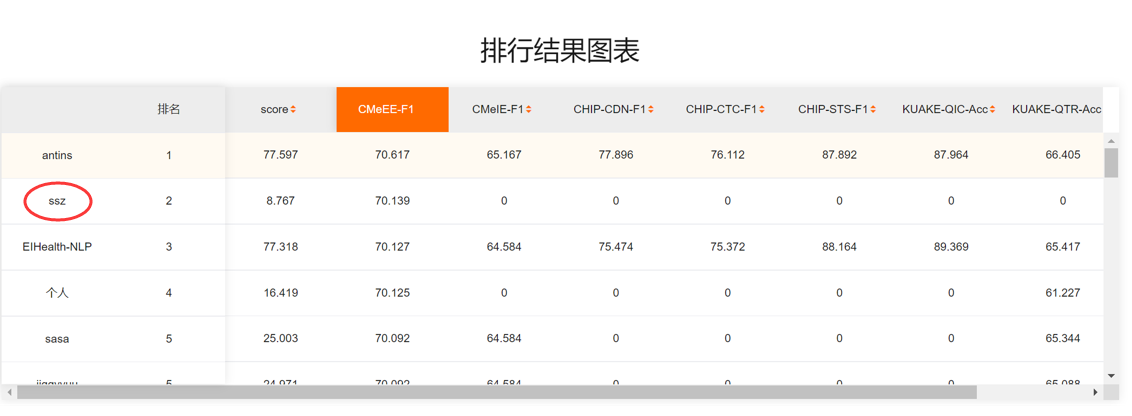
We got the 2nd price of the benchmark by 2022/06/13.
Single model CBioNAMER has surpassed human(67.0 in F1-score).
Results of our method:
Results of our single model CBioNAMER:
CBioNAMER is a sub-model in our result, which is based on GlobalPointer (a powerful open-source model, we rewrite it with Pytorch) and MacBert.
First, install PyTorch>=1.7.0. There's no restriction on GPU or CUDA.
Then, install this repo as a Python package:
$ pip install CBioNAMERPython package transformers==4.6.1 would be automatically installed as well.
The CBioNAMER package provides the following methods:
CBioNAMER.load_NER(model_save_path='./checkpoint/macbert-large_dict.pth', maxlen=512, c_size=9, id2c=_id2c, c2c=_c2c)
Returns the pretrained model. It will download the model as necessary. The model would use the first CUDA device if there's any, otherwise using CPU instead.
The model_save_path argument specifies the path of the pretrained model weight.
The maxlen argument specifies the max length of input sentences. The sentences longer than maxlen would be cut off.
The c_size argument specifies the number of entity class. Here is 9 for CBLUE.
The id2c argument specifies the mapping between id and entity class. By default, the id2c argument for CBLUE is:
_id2c = {0: 'dis', 1: 'sym', 2: 'pro', 3: 'equ', 4: 'dru', 5: 'ite', 6: 'bod', 7: 'dep', 8: 'mic'}
The c2c argument specifies the mapping between entity class and its Chinese meaning. By default, the c2c argument for CBLUE is:
_c2c = {'dis': "疾病", 'sym': "临床表现", 'pro': "医疗程序", 'equ': "医疗设备", 'dru': "药物", 'ite': "医学检验项目", 'bod': "身体", 'dep': "科室", 'mic': "微生物类"}
The model returned by CBioNAMER.load_NER() supports the following methods:
model.recognize(text: str, threshold=0)
Given a sentence, returns a list of dictionaries with recognized entity, the format of the dictionary is {'start_idx': entity's starting index, 'end_idx': entity's ending index, 'type': entity class, 'Chinese_type': Chinese meaning of entity class, 'entity': recognized entity}. The threshold argument specifies that the returned list only contains the recognized entity with confidence score higher than threshold.
model.predict_to_file(in_file: str, out_file: str)
Given input and output .json file path, the model would do inference according in_file, and the recognized entity would be saved in out_file. The output file can be submitted to CBLUE. The format of input file is like:
[
{
"text": "该技术的应用使某些遗传病的诊治水平得到显著提高。"
},
...
{
"text": "There is a sentence."
}
]import CBioNAMER
NER = CBioNAMER.load_NER()
in_file = './CMeEE_test.json'
out_file = './CMeEE_test_answer.json'
NER.predict_to_file(in_file, out_file)import CBioNAMER
NER = CBioNAMER.load_NER()
text = "该技术的应用使某些遗传病的诊治水平得到显著提高。"
recognized_entity = NER.recognize(text)
print(recognized_entity)
# output:[{'start_idx': 9, 'end_idx': 11, 'type': 'dis', 'Chinese_type': '疾病', 'entity': '遗传病'}]