What is MEA-NAP? | Documentation | Citing MEA-NAP | Features | Installation | How to use the pipeline | Video tutorials | Example dataset | Troubleshooting | Contributing
Alterations in synaptic function, and other cellular processes that affect neuronal communication, can alter the trajectory of network development in neuronal circuits. New tools are needed for studying network function at the cellular-scale that are accessible to cellular neuroscientists and stem cell biologists recording microelectrode array (MEA) data from 2D and 3D rodent or human-derived neuronal cultures. Our microelectrode array (MEA) network analysis pipeline (MEA-NAP) combines methods for studying network function using graph theoretical and other network metrics that are commonly applied at the whole brain level (e.g., fMRI data) and in other fields of network science. The aim of the pipeline is to facilitate comparisons of microscale network function from MEA recordings including functional connectivity, network topology, and network dynamics. These network features facilitate comparison of information processing and computational efficiency. Thus, microscale network function can provide a platform for testing new therapeutic strategies, including pharmacologic therapies and stimulating specific nodes in the network to modulate network function, to address information processing deficits in neurologic disease models.
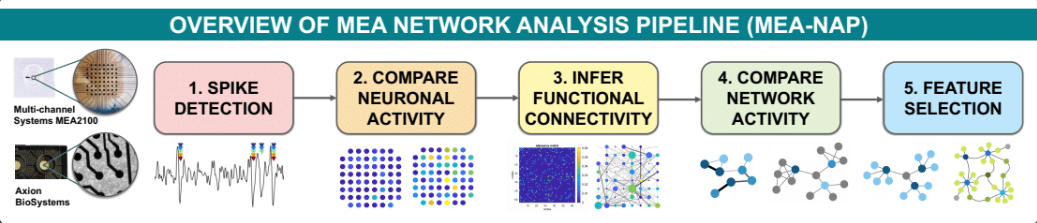
MEA-NAP is a streamlined diagnostic and analytic tool for microscale network activity data obtained using microelectrode arrays. MEA-NAP provides a straight forward way for new and experienced MATLAB users to quickly compare spike detection methods, neuronal activity (including firing rate and burst detection), functional connectivity, network topology (including network metrics from graph theory), and network dynamics. MEA-NAP performs batch analysis of an experimental dataset (e.g., MEA recordings from wild-type and knock-out cultures at multiple developmental time points). MEA-NAP produces summary plots and performs statistics on these features and organizes the output figures in a convenient file structure. The user can then identify network-level developmental or genotypic differences in their MEA dataset. The pipeline is written in MATLAB and was designed for experimentalists with little or no experience with network analysis. Experienced users will find the batch analysis and automatic figure generation convenient for examining both individual network and group comparisons.
Please see our detailed documentation for MEA-NAP users at our read-the-docs page.
You can view our video tutorials at YouTube or download the MEA-NAP GUI tutorial video at the Harvard Dataverse.
Our manuscript describing MEA-NAP is now available as a pre-print on bioRxiv!
Timothy PH Sit, Rachael C Feord, Alexander WE Dunn, Jeremi Chabros, David Oluigbo, Hugo H Smith, Lance Burn, Elise Chang, Alessio Boschi, Yin Yuan, George M Gibbons, Mahsa Khayat-Khoei, Francesco De Angelis, Erik Hemberg, Martin Hemberg, Madeline A Lancaster, Andras Lakatos, Stephen J Eglen, Ole Paulsen, Susanna B Mierau. MEA-NAP compares microscale functional connectivity, topology, and network dynamics in organoid or monolayer neuronal cultures. bioRxiv 2024.02.05.578738. doi: https://doi.org/10.1101/2024.02.05.578738
MEA-NAP compares metrics for functional connectivity, network topology, and network dynamics. This includes many graph theoretical metrics from the Brain Connectivity Toolbox (commonly applied to study networks at the whole brain macro scale) to microscale functional networks from MEA recordings of neuronal cultures or brain organoid slices. This is achieved using cutting-edge methods for determining significant functional connections including the spike time tiling coefficient and probablistic thresholding. MEA-NAP also includes state-of-the-art normalization techniques for graph metrics to facilitate comparison of different sized networks. MEA-NAP includes the first implementation at the microscale of node cartography, for classifying nodal roles within the network, and effective rank, for calculating the number of subcommunities within the microscale networks based on their activity patterns. New features also include control theoretical metrics to identify nodes that can drive network activity and dimensionality reduction using non-negative matrix factorization to characterize patterns of activity observed in the network. Applications to 3D human cerebral organoid, 2D human iPSC-derived and mouse neuronal cultures are highlighted in our MEA-NAP manuscript.
Importantly, MEA-NAP analyses the MEA experiments through batch analysis automatically adjusting scaling and production of comparison plots to the whole dataset. This allows users new to network neuroscience to identify the most interesting network-level features in their dataset to explore further. The plots can also be exported as png (e.g., for adding to presentations or posters) or svg (e.g., for adjusting font or color in Illustrator or Affinity Designer).
Learn more about the network features and validation tools in MEA-NAP in the full documentation.
You can see our MEA-NAP FENS poster we presented at the 2022 Federation of European Neuroscience Societies (FENS) Forum.
For new users, please see Setting up MEA-NAP.
For experienced users, clone the github repository to a location of your choice (e.g., your desktop folder):
git clone https://github.com/SAND-Lab/MEA-NAP
To confirm you have the appropriate version of Matlab and Matlab toolboxes installed, please see the MEA-NAP systems requirments. Our detailed documentation for MEA-NAP provides step-by-step instructions for formatting your data and running MEA-NAP.
To get the cutting edge of the pipeline, which includes the latest features / bug fixes but may introduce new errors, do:
git clone https://github.com/SAND-Lab/MEA-NAP
git checkout dev
To quickly get started, open MEApipeline.m in matlab.
You will first need to ensure that your data has been converted to mat files with the appropriate variables and that you have created a spreadsheet (csv file) with the names of your mat files for each recording and their group and ages to guide the batch analysis.
If your data is in the right format, you can then press run in MEApipeline.m. The guided user interface (GUI) will prompt you to select the location of the folder where you downloaded MEA-NAP, the folder with your data, and the name and location of the batch analysis csv or xlsx file. Then the batch analysis will run autonomously.
New users can watch our video tutorial at https://www.youtube.com/watch?v=oxFyqRyemRM or download the video at the Harvard Dataverse https://doi.org/10.7910/DVN/Z14LWA.
For advanced MATLAB users, in MEApipeline.m in matlab, you can read through the instructions to customize your choice of parameters. Many of these parameters can also be changed in the GUI by selecting "Show Advanced Settings."
You can find the full documentation on our MEA-NAP read-the-docs website.
An example MEA dataset with the corresponding batch analysis file is available at the Harvard Dataverse https://doi.org/10.7910/DVN/Z14LWA. The output folder including all the analysis and figures generated by MEA-NAP from this dataset is also available at the Dataverse.
If you experience an error whilst running MEA-NAP, please download MEA-NAP again from our Github webpage. We routinely update MEA-NAP as we add new features, solve errors created by edge cases, or fix other issues.
For most issues, please use the Issues tab on github and open a new issue, e.g., see an example issue here.
For SAND Lab users or collaborators: If it is urgent or requires long discussion with multiple lab members, you can also send a message on Slack or email.
We are always keen to have collaborators who are interested in contributing to the application of network metrics and/or code in MEA-NAP. Please let us know if you are interested.
For current SAND users and collaborators, please see contributing code instructions in the MEA-NAP documentation.
The documentation of this toolbox is managed using read-the-docs and sphinx. To build and test the website on your local computer, you first need a python environment (eg. see anaconda), then you want to install sphinx
pip install sphinx
and some extra packages used in this documentation
pip install sphinx-hoverxref
and also install the theme used for the documentation website:
pip install furo
then navigate to the folder containing the documentation, which should be in /your/path/to/AnalysisPipeline/docs/, then do:
make html
you can then open your website in _build/html/index.html, and read the "How to contribute" section of the website to learn more about editing the documentation.