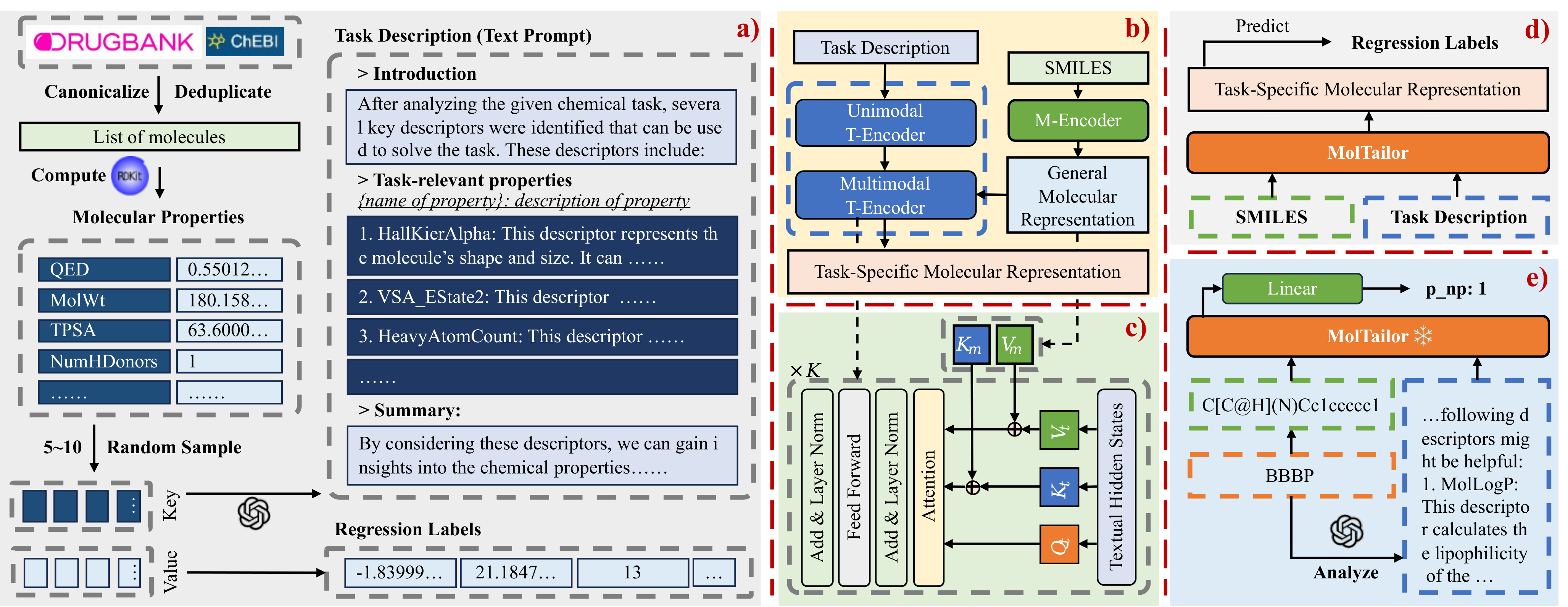
NOTE: In this project, MolTailor is referred to as DEN.
Deep learning is now widely used in drug discovery, providing significant acceleration and cost reduction. As the most fundamental building block, molecular representation is essential for predicting molecular properties to enable various downstream applications. Most existing methods attempt to incorporate more information to learn better representations. However, not all features are equally important for a specific task. Ignoring this would potentially compromise the training efficiency and predictive accuracy. To address this issue, we propose a novel approach, which treats language models as an agent and molecular pretraining models as a knowledge base. The agent accentuates task-relevant features in the molecular representation by understanding the natural language description of the task, just as a tailor customizes clothes for clients. Thus, we call this approach MolTailor. Evaluations demonstrate MolTailor's superior performance over baselines, validating the efficacy of enhancing relevance for molecular representation learning. This illustrates the potential of language model guided optimization to better exploit and unleash the capabilities of existing powerful molecular representation methods.
.
├── mt-mtr-build # Code for building the MT-MTR pre-training corpus
│ ├── 01-get-smiles.py # Merge SMILES from DrugBank and ChEBI
│ ├── 02-calculate-descriptors.py # Calculate molecular descriptors using RDKit
│ ├── 03-generate-descriptions.py # Generate task descriptions using GPT-3.5
│ ├── 04-build-mt-mtr.py # Build the MT-MTR corpus
│ └── data
│ ├── temporary # Store intermediate data
│ ├── mt-mtr.pt # MT-MTR
│ ├── mt-mtr-clean.pt # MT-MTR with molecules duplicated in 8 MoleculeNet datasets removed
│ ├── mt-mtr-origin.jsonl # MT-MTR with all molecular properties
│ └── mt-mtr-origin-clean.jsonl
├── pretrain # Pre-training code
│ ├── data # Store pre-training corpus mt-mtr.pt
│ ├── workspace # Checkpoint and log files
│ ├── models # Model structures
│ │ ├── bert_uncased # Backbone model code
│ │ ├── ...
│ │ ├── bert.py # Construct Multimodal T-Encoder based on BERT code
│ │ ├── config.py # MolTailor configuration file
│ │ ├── den.py # Specific implementation of MolTailor
│ │ ├── load.py # Load backbone
│ │ └── multitask.py # Pre-training task code
│ ├── data_collator.py
│ ├── data_modules.py
│ ├── dataset.py
│ ├── debug.py
│ ├── main.py # Entry file
│ ├── train.py
│ └── tune.py # Search for batch size and learning rate
├── linear-probe-moleculenet # Downstream task code
│ ├── models # Code for MolTailor and Baseline models
│ ├── data # MoleculeNet dataset and related preprocessing code
│ │ ├── feature # Extracted molecular embeddings
│ │ ├── raw # Original MoleculeNet dataset
│ │ ├── utils
│ │ │ ├── preprocess.py # MoleculeNet preprocessing code
│ │ │ └── feature-extract.py # Extract molecular embeddings
│ │ ├── prompt4molnet.json # Text prompts for MolTailor in downstream tasks
│ │ ├── bbbp.csv # Preprocessed MoleculeNet dataset
│ │ └── ...
│ ├── workspace # Checkpoint and log files
│ ├── callbacks.py # Callback functions for lightning
│ ├── data_modules.py
│ ├── dataset.py
│ ├── main.py # Entry file
│ ├── metrics.py # Implementation of roc_auc and delta ap methods
│ ├── multi_seeds.py
│ ├── split.py # Random and scaffold dataset splitting function implementations
│ ├── train.py
│ └── tune.py # Search for learning rate using optuna
├── linear-probe-moleculenet-lite # Downstream task code without Uni-Mol model, file structure is the same as above
│ └── ...
├── models # 模型权重
│ ├── BiomedNLP-PubMedBERT-base-uncased-abstract-fulltext # Default text backbone
│ ├── CHEM-BERT # Default molecular backbone
│ ├── ChemBERTa-10M-MTR
│ ├── DEN # MolTailor is named DEN in this project
│ │ ├── 0al3aezz # Using BioLinkBERT and ChemBERTa as backbones
│ │ ├── u02pzsl2 # Using PubMedBERT and ChemBERTa as backbones
│ │ ├── f9x97q2q # Using PubMedBERT and CHEM-BERT as backbones
│ │ └── ...
│ └── ...
├── readme
├── readme.md
├── requirements-lite.txt # Dependencies without Uni-Mol model
├── requirements.txt # Full dependencies
└── scripts
├── pretrain.sh # Pre-training script
├── convert_ckpt.sh # Move and convert the pre-trained model to the models folder
├── linear-probe-molnet.sh # Downstream task script
└── linear-probe-molnet-lite.shDue to the complex environment setup required for the Uni-Mol baseline model in downstream tasks, we offer two environment configuration options: a full version and a lightweight version. The full version has strict CUDA version requirements due to dependencies on the Uni-Mol model. If your environment doesn't meet these requirements, you may opt for the lightweight configuration.
conda create -n moltailor python=3.9
conda activate moltailor
# It is recommended to use the specified versions of PyTorch and CUDA here to meet the dependencies of the uni-mol model later on.
conda install pytorch==2.0.0 torchvision==0.15.0 torchaudio==2.0.0 pytorch-cuda=11.8 -c pytorch -c nvidia
cd MolTailor/
pip install -r requirements.txt
pip install dgl -f https://data.dgl.ai/wheels/cu118/repo.html
pip install torch-scatter -f https://data.pyg.org/whl/torch-2.0.0+cu118.html
# Download Uni-Core wheel. You can download the version suitable for your system from the original project: https://github.com/dptech-corp/Uni-Core/releases/tag/0.0.3
wget https://github.com/dptech-corp/Uni-Core/releases/download/0.0.3/unicore-0.0.1+cu118torch2.0.0-cp39-cp39-linux_x86_64.whl
pip install unicore-0.0.1+cu118torch2.0.0-cp39-cp39-linux_x86_64.whl
# Delete the wheel file after installation
rm unicore-0.0.1+cu118torch2.0.0-cp39-cp39-linux_x86_64.whlconda create -n moltailor python=3.9
conda activate moltailor
# You can choose versions that suit your system
conda install pytorch==2.0.0 torchvision==0.15.0 torchaudio==2.0.0 pytorch-cuda=11.8 -c pytorch -c nvidia
cd MolTailor/
pip install -r requirements.txt
# You can download the version that suits your system from the official website: https://www.dgl.ai/pages/start.html
pip install dgl -f https://data.dgl.ai/wheels/cu118/repo.html
# You can download the version that suits your system from the official website: https://pypi.org/project/torch-scatter/
pip install torch-scatter -f https://data.pyg.org/whl/torch-2.0.0+cu118.htmlYou can download the dataset from Google Drive or Baidu Netdisk. All data files in the project are stored in the compressed file MolTailor-Data.zip following their original directory structure. Simply download and unzip this file, then move the data folder to the corresponding directory.
If you are only interested in the MT-MTR dataset, you can download the MT-MTR.zip file. After unzipping, the meaning of each file can be found in the comments of the subsection File Structure.
You can download the model weights from Google Drive or Baidu Netdisk. The MolTailor-Models folder contains weights for MolTailor and models that cannot be directly downloaded from Huggingface. Download the necessary model zip files, unzip them, and then move the folders to the MolTailor/models/ directory.
For pre-training, the training process requires loading text and molecular modality backbones, which by default are PubMedBERT and CHEM-BERT. Therefore, you need to use the following commands to download the corresponding weight files.
For downstream tasks, you need to download the weight files corresponding to the model you want to test and move them to the MolTailor/models/ folder after unzipping. It is important to note that the weight file for the MolTailor model is DEN.zip. Specifically, we provide three versions of MolTailor, which are:
- 0al3aezz: Uses BioLinkBERT and ChemBERTa as backbones
- u02pzsl2: Uses PubMedBERT and ChemBERTa as backbones
- f9x97q2q: Uses PubMedBERT and CHEM-BERT as backbones
The following models can be downloaded from Huggingface and are not included in the MolTailor-Models folder:
git lfs install
cd MolTailor/models
# BERT
git clone https://huggingface.co/google-bert/bert-base-uncased
# RoBERTa
git clone https://huggingface.co/FacebookAI/roberta-base
# SciBERT
git clone https://huggingface.co/allenai/scibert_scivocab_uncased
# BioLinkBERT
git clone https://huggingface.co/michiyasunaga/BioLinkBERT-base
# PubMedBERT
git clone https://huggingface.co/microsoft/BiomedNLP-BiomedBERT-base-uncased-abstract-fulltext
# ChemBERTa-10M-MTR
git clone https://huggingface.co/DeepChem/ChemBERTa-10M-MTR
# ChemBERTa-77M-MLM
git clone https://huggingface.co/DeepChem/ChemBERTa-77M-MLM
# ChemBERTa-77M-MTR
git clone https://huggingface.co/DeepChem/ChemBERTa-77M-MTR
# T5
git clone https://huggingface.co/google-t5/t5-base
# MolT5
git clone https://huggingface.co/laituan245/molt5-base
# TCT5
git clone https://huggingface.co/GT4SD/multitask-text-and-chemistry-t5-base-augmPlease first refer to the instructions in the section Data and Weights to download the necessary data and weight files.
If you are interested in the process of constructing the MT-MTR corpus or wish to build your own MT-MTR corpus, you can refer to the code in the mt-mtr-build folder.
You can execute the following commands to start pre-training:
cd scripts
zsh pretrain.shIt's important to note that our hyperparameters for pre-training were set based on our hardware resources (two A100-80G GPUs). You may need to adjust the hyperparameters based on your own resources.
Also, our provided pre-training code only supports BERT-like models such as PubMedBERT and BioLinkBERT as the text backbone, and CHEM-BERT as the molecular backbone.
The pre-training code for using ChemBERTa as the molecular backbone is not provided, but the corresponding model weights are available for testing in downstream tasks. If you are interested in pre-training with ChemBERTa as the molecular backbone, you can raise an issue, and we will provide the corresponding code in the future.
After pre-training, you can execute the following script to move the model weights to the MolTailor/models/ folder for subsequent downstream tasks:
cd scripts
zsh convert_ckpt.shPlease ensure that there is a DEN folder under MolTailor/models/ before proceeding.
Please first refer to the instructions in the section Data and Weights to download the necessary data and weight files.
We have selected 8 tasks from MoleculeNet for downstream tasks, namely: BBBP, ClinTox, HIV, Tox21, ESOL, FreeSolv, Lipophilicity, and QM8. You can run the downstream task code by executing the following commands:
cd scripts
# If you have set up the full environment
zsh linear-probe-molnet.sh MODEL_NAME
# If you have set up the lightweight environment
zsh linear-probe-molnet-lite.sh MODEL_NAMESupported model names include:
Random、RDKit-FP、Morgan-FP、MACCS-FP、RDKit-DP、KCL、Grover、MolCLR、MoMu、CLAMP、Uni-Mol、Mole-BERT、CHEM-BERT、BERT、RoBERTa、SciBERT、PubMedBERT、BioLinkBERT、ChemBERTa-77M-MTR、ChemBERTa-10M-MTR、ChemBERTa-77M-MLM、MolT5、T5、TCT5、DEN-f9x97q2q、DEN-ChemBERTa-u02pzsl2、DEN-ChemBERTa-0al3aezz、
Among these, DEN-f9x97q2q represents MolTailor using PubMedBERT and CHEM-BERT as backbones, DEN-ChemBERTa-u02pzsl2 represents MolTailor using PubMedBERT and ChemBERTa as backbones, and DEN-ChemBERTa-0al3aezz represents MolTailor using BioLinkBERT and ChemBERTa as backbones.
@article{guo2024moltailor,
title={MolTailor: Tailoring Chemical Molecular Representation to Specific Tasks via Text Prompts},
author={Guo, Haoqiang and Zhao, Sendong and Wang, Haochun and Du, Yanrui and Qin, Bing},
journal={arXiv preprint arXiv:2401.11403},
year={2024}
}