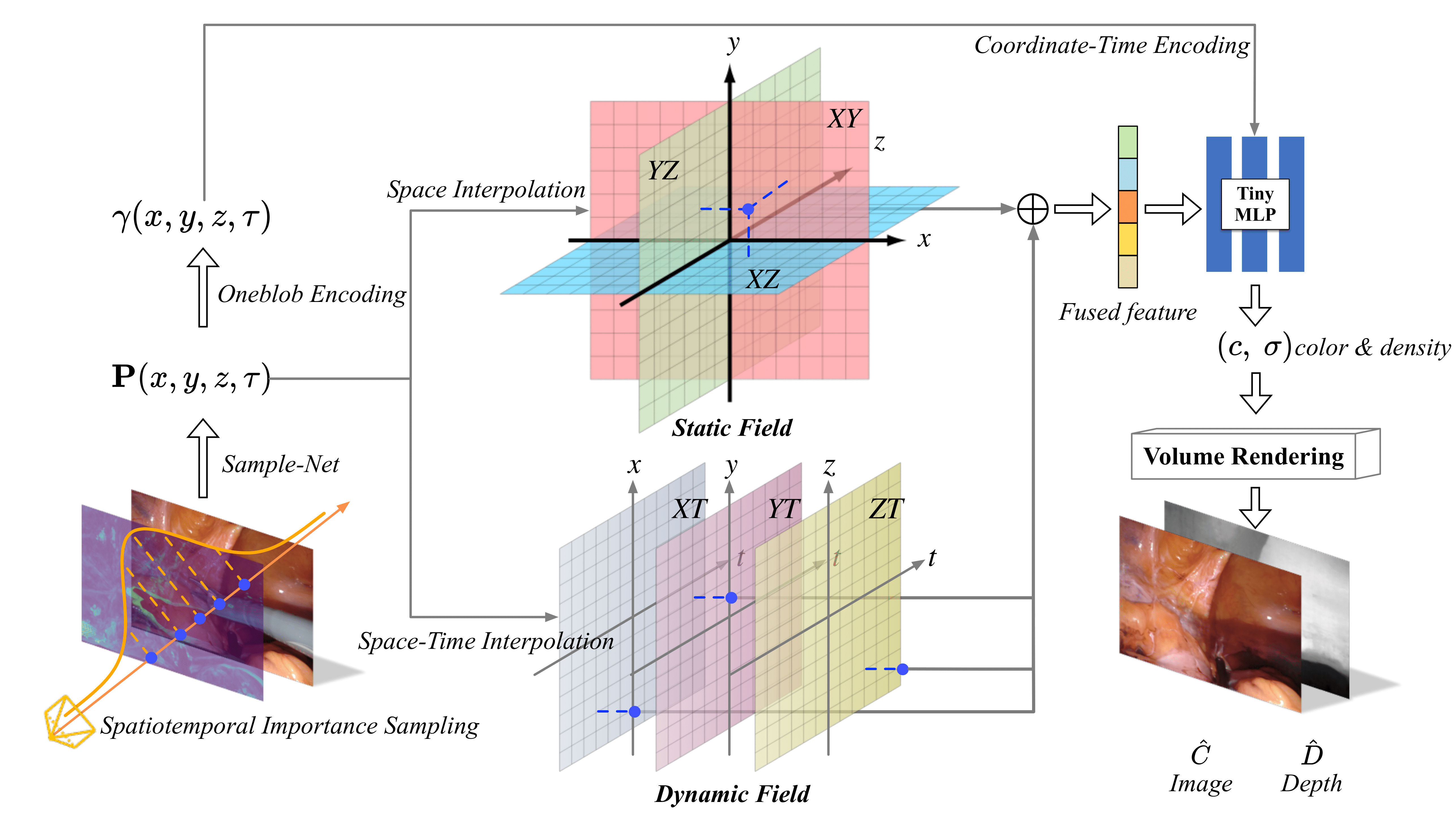
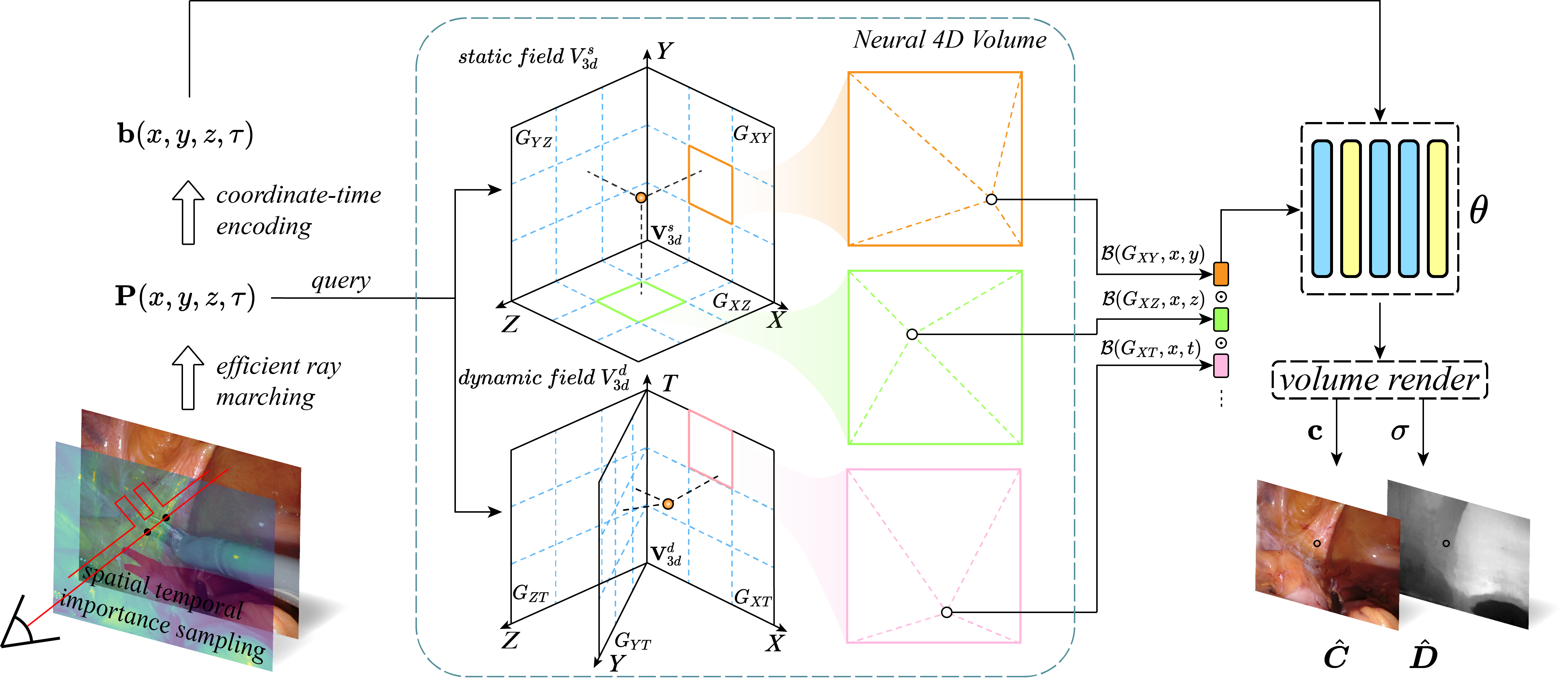
LerPlane: Neural Representations for Fast 4D Reconstruction of Deformable Tissues
Chen Yang, Kailing Wang, Yuehao Wang, Xiaokang Yang, Wei Shen
MICCAI2023, Young Scientist Award
Efficient Deformable Tissue Reconstruction via Orthogonal Neural Plane
Chen Yang, Kailing Wang, Yuehao Wang, Qi Dou, Xiaokang Yang, Wei Shen
TMI2024
- Initial Code Release.
- Further check of the reproducibility.
- Code Refactoring and Realease of the final version.
- Hamlyn Dataset.
Reconstructing deformable tissues from endoscopic stereo videos in robotic surgery is crucial for various clinical applications. However, existing methods relying only on implicit representations are computationally expensive and require dozens of hours, which limits further practical applications. To address this challenge, we introduce LerPlane, a novel method for fast and accurate reconstruction of surgical scenes under a single-viewpoint setting. LerPlane treats surgical procedures as 4D volumes and factorizes them into explicit 2D planes of static and dynamic fields, leading to a compact memory footprint and significantly accelerated optimization. The efficient factorization is accomplished by fusing features obtained through linear interpolation of each plane and enabling the use of lightweight neural networks to model surgical scenes. Besides, LerPlane shares static fields, significantly reducing the workload of dynamic tissue modeling. We also propose a novel sample scheme to boost optimization and improve performance in regions with tool occlusion and large motions. Experiments on DaVinci robotic surgery videos demonstrate that LerPlane accelerates optimization by over 100× while maintaining high quality across various non-rigid deformations, showing significant promise for future intraoperative surgery applications.
trainging_speed_vs_endo.mp4
Tested with an Ubuntu workstation i9-12900K, 3090GPU.
conda create -n lerplane python=3.9
conda activate lerplane
pip install -r requirements.txt
pip install git+https://github.com/NVlabs/tiny-cuda-nn/#subdirectory=bindings/torch
We notice tiny-cuda-nn is sometimes not compilable on some latest GPUs like RTX4090(tested 2023.1). If you found OSError while installing tiny-cuda-nn, you can refer to this issue or this article. We've successfully built the same env on an Ubuntu 22.04 workstation i7-13700K, 4090GPU with the commands above.
Download the datasets
Please download the dataset from EndoNeRF
To use the example config, organize your data like:
data
| - endonerf_full_datasets
| | - cutting_tissues_twice
| | - pushing_soft_tissues
| - hamlyn_forplane
| | - hamlyn1
| | - hamlyn2
| - YourCustomDatasets
Using configs for training
Lerplane uses configs to control the training process. The example configs are stored in the lerplanes/config folder.
To train a model, run the following command:
export CUDA_VISIBLE_DEVICES=0
PYTHONPATH='.' python lerplanes/main.py --config-path lerplanes/config/example-9k.py
We use the same evaluation protocol as EndoNeRF. So please follow the instructions in EndoNeRF.
We would like to acknowledge the following inspiring work:
Big thanks to NeRFAcc (Li et al.) for their efficient implementation, which has significantly accelerated our rendering.
If you find this code useful for your research, please use the following BibTeX entries:
@inproceedings{yang2023neural,
title={Neural lerplane representations for fast 4d reconstruction of deformable tissues},
author={Yang, Chen and Wang, Kailing and Wang, Yuehao and Yang, Xiaokang and Shen, Wei},
booktitle={International Conference on Medical Image Computing and Computer-Assisted Intervention},
pages={46--56},
year={2023},
organization={Springer}
}
and
@article{yang2024efficient,
title={Efficient deformable tissue reconstruction via orthogonal neural plane},
author={Yang, Chen and Wang, Kailing and Wang, Yuehao and Dou, Qi and Yang, Xiaokang and Shen, Wei},
journal={IEEE Transactions on Medical Imaging},
year={2024},
publisher={IEEE}
}