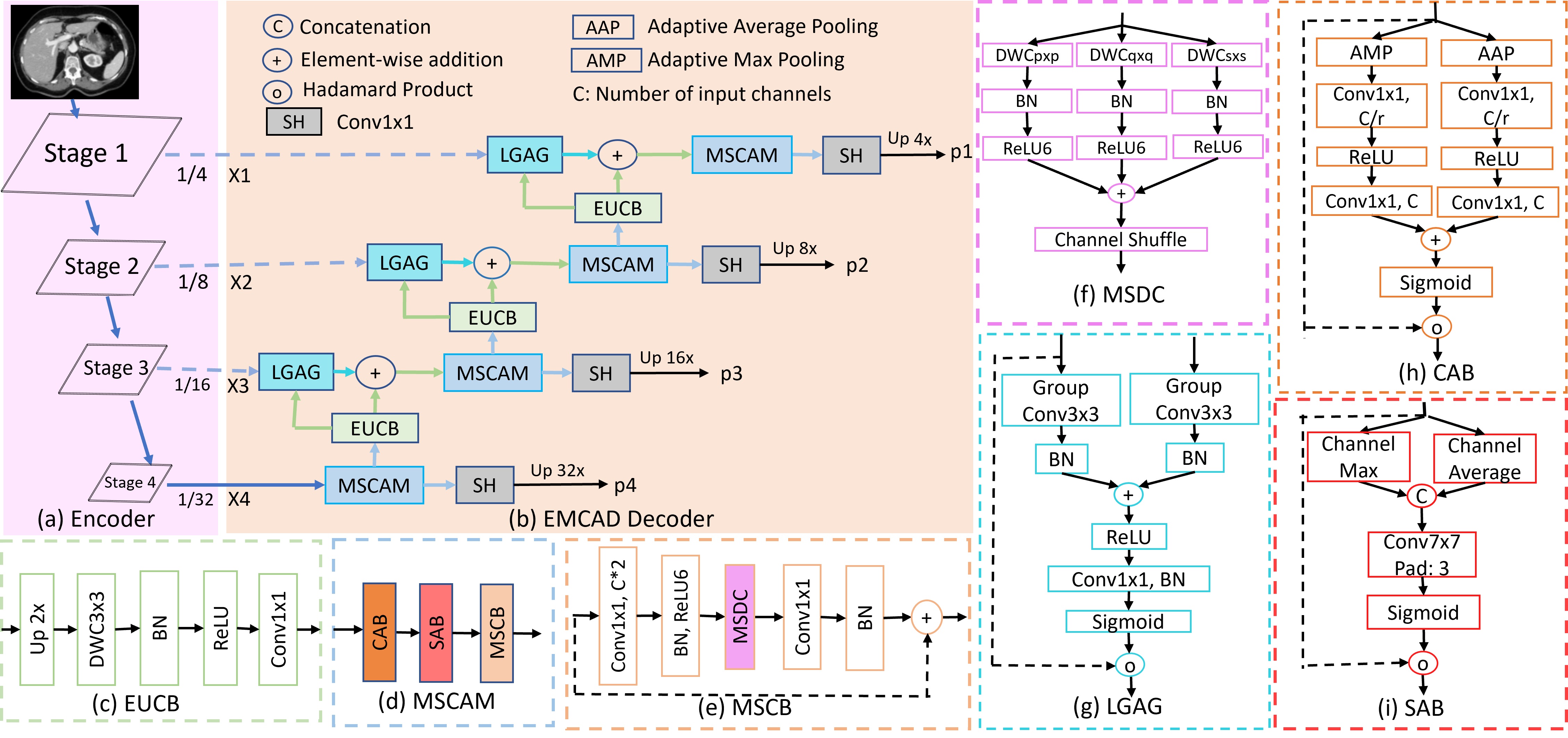
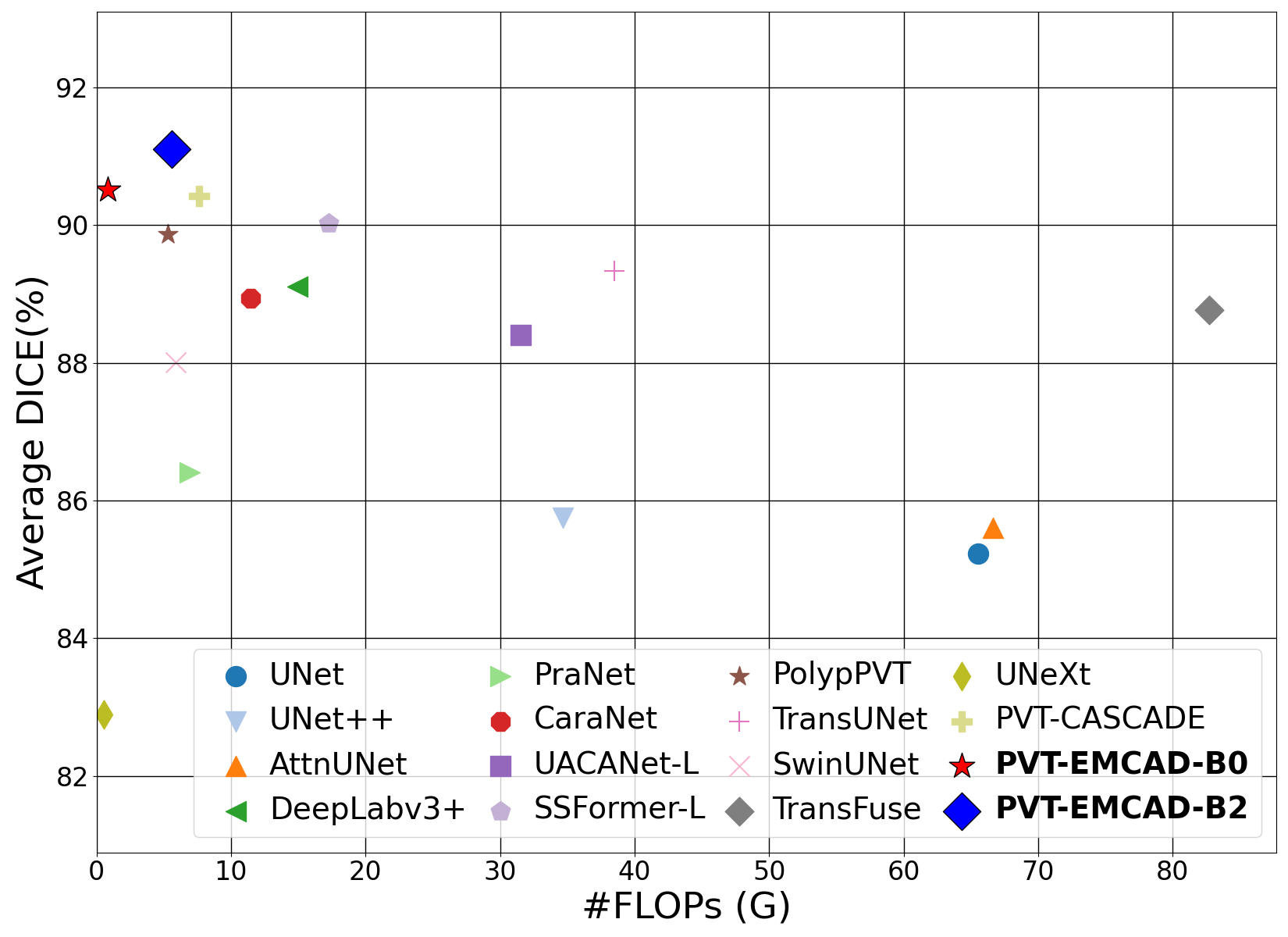
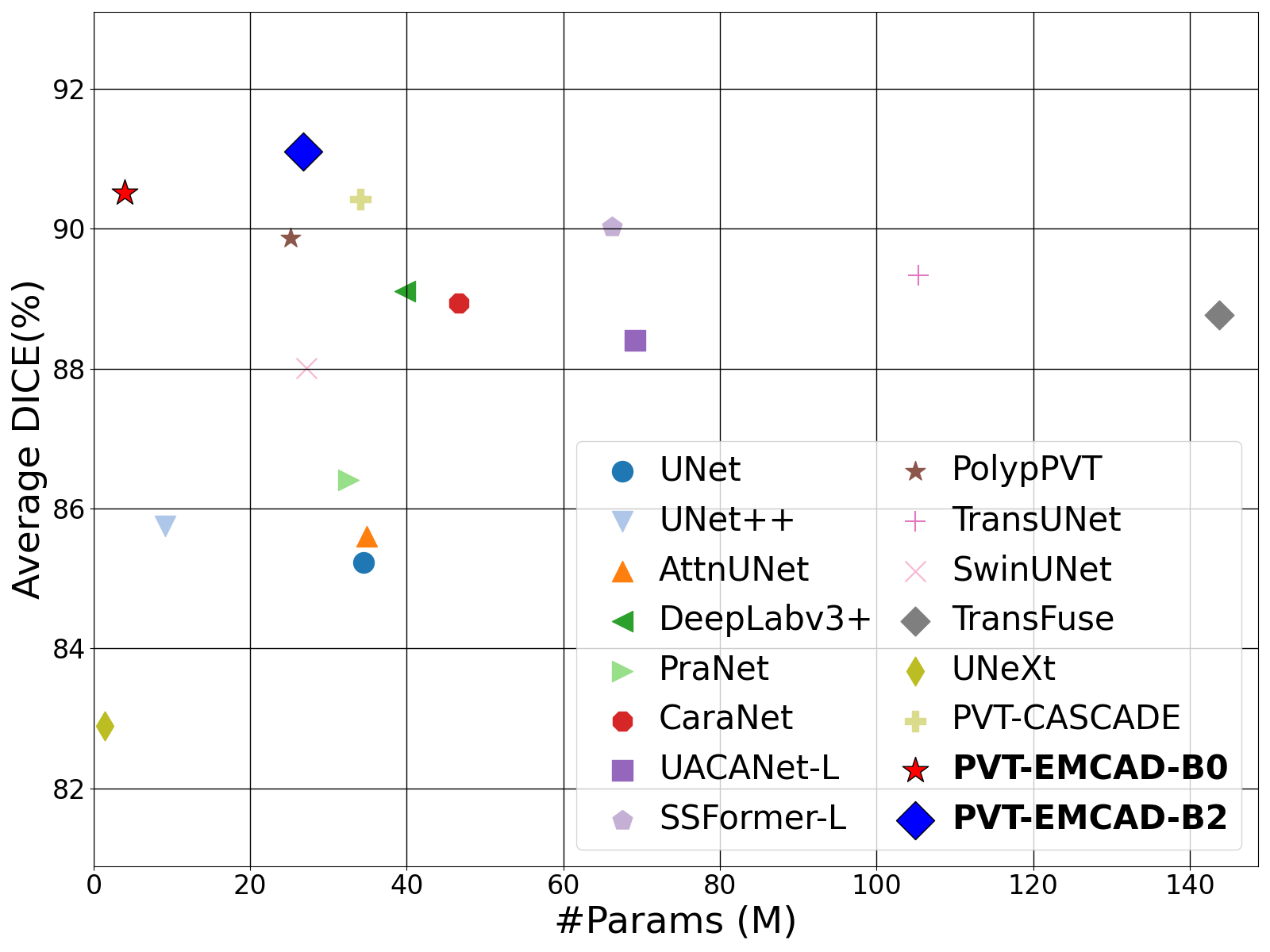
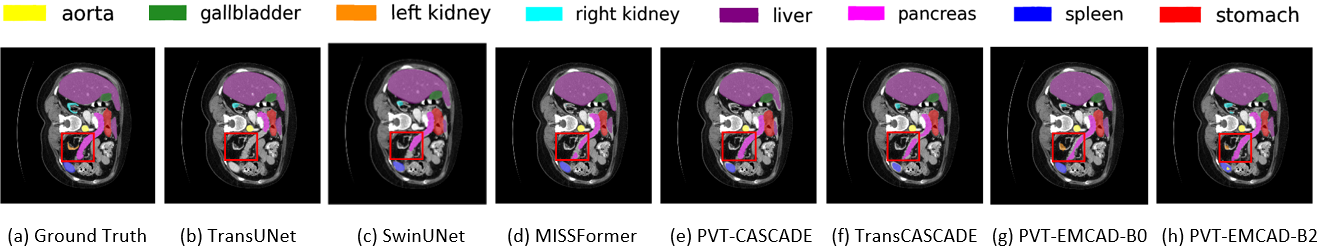
Official Pytorch implementation of the paper EMCAD: Efficient Multi-scale Convolutional Attention Decoding for Medical Image Segmentation published in CVPR 2024. arxiv code video
Md Mostafijur Rahman, Mustafa Munir, Radu Marculescu
The University of Texas at Austin
Synapse training code released!!!
Please run the following commands.
conda create -n emcadenv python=3.8
conda activate emcadenv
pip install torch==1.11.0+cu113 torchvision==0.12.0+cu113 torchaudio==0.11.0 --extra-index-url https://download.pytorch.org/whl/cu113
pip install mmcv-full -f https://download.openmmlab.com/mmcv/dist/cu113/torch1.11.0/index.html
pip install -r requirements.txt
-
Synapse Multi-organ dataset: Sign up in the official Synapse website and download the dataset. Then split the 'RawData' folder into 'TrainSet' (18 scans) and 'TestSet' (12 scans) following the TransUNet's lists and put in the './data/synapse/Abdomen/RawData/' folder. Finally, preprocess using
python ./utils/preprocess_synapse_data.pyor download the preprocessed data and save in the './data/synapse/' folder. Note: If you use the preprocessed data from TransUNet, please make necessary changes (i.e., remove the code segment (line# 88-94) to convert groundtruth labels from 14 to 9 classes) in the utils/dataset_synapse.py. -
ACDC dataset: Download the preprocessed ACDC dataset from Google Drive of MT-UNet and move into './data/ACDC/' folder.
You should download the pretrained PVTv2 model from Google Drive/PVT GitHub, and then put it in the './pretrained_pth/pvt/' folder for initialization.
cd into EMCAD
python -W ignore train_synapse.py --root_path /path/to/train/data --volume_path path/to/test/data --encoder pvt_v2_b2 # replace --root_path and --volume_path with your actual path to data.
cd into EMCAD
We are very grateful for these excellent works timm, CASCADE, MERIT, G-CASCADE, PraNet, Polyp-PVT and TransUNet, which have provided the basis for our framework.
@inproceedings{rahman2024emcad,
title={Emcad: Efficient multi-scale convolutional attention decoding for medical image segmentation},
author={Rahman, Md Mostafijur and Munir, Mustafa and Marculescu, Radu},
booktitle={Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition},
pages={11769--11779},
year={2024}
}