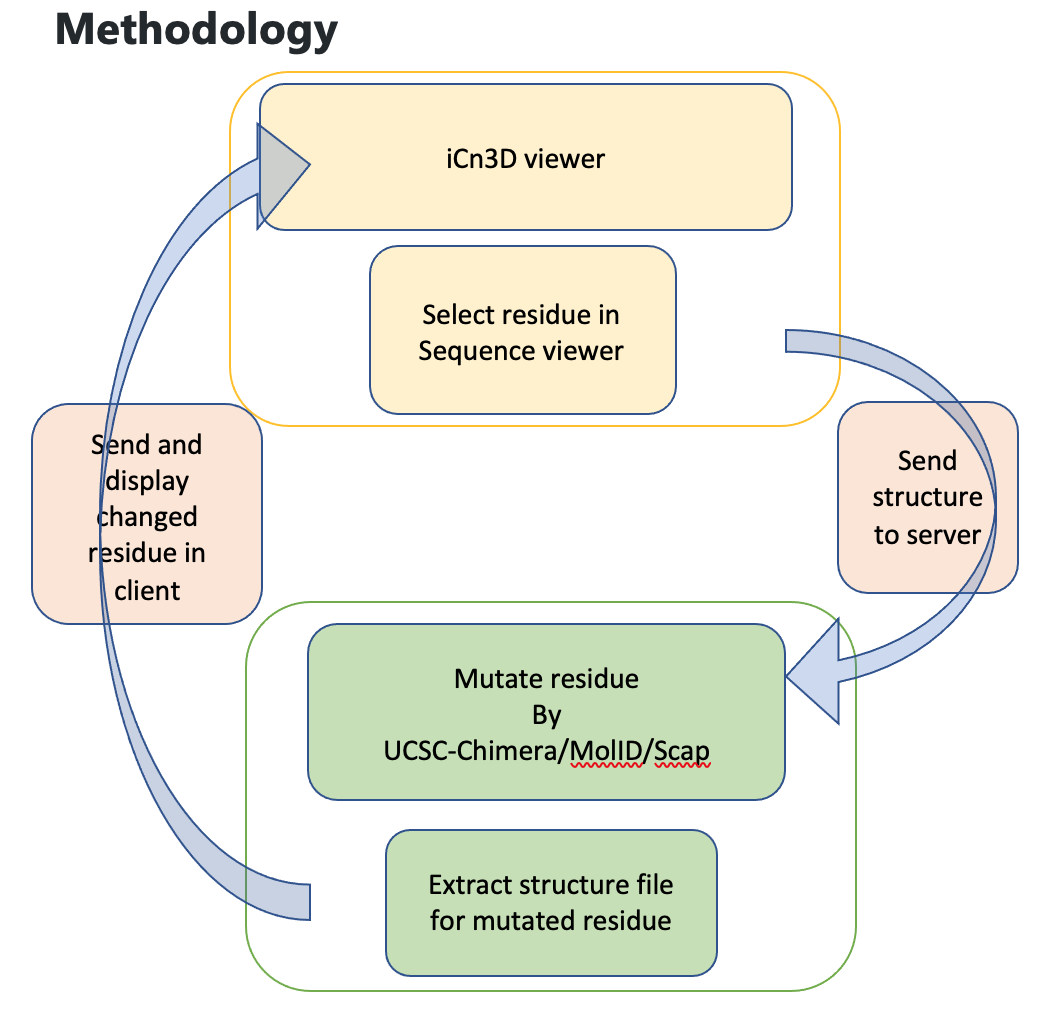
iCn3D provides a web-based tool for protein structure visualizing. Here we seek to expand the functionality of this tool to:
- integrate 3D visualization of SNPs
- Build on the functioning SymD integration to include local symmetry analysis capabilities
There are several tools available for visualizing structural homology modelling (SCWRL, AMBER), most are not available in a web based format, or are not licensed outside for use outside of academia. We explored integration of both scap (http://honig.c2b2.columbia.edu/jackal), chimera based scripts, and modeller to show mutated residues in 3D.
- Modeller's mutate_model.py script was used to do the following mutations in 1TUP (pdb1tup.ent)
- L111->PRO in Chain A (1tupPRO111.pdb)
- F113->VAL in Chain B (1tupVAL113.pdb)
- Modeller package
- using script at github repo https://github.com/sridharacharya/bioStructureTools
- Usage: Structure.py -i <pdb-id> or -f <pdb-file> -r <included resno> -x <excluded resno> -o <output-file-name>
python Structure.py -f 1tupPRO111.pdb -r A111 -o 1tupPRO111_only.pdb
python Structure.py -f 1tupVAL113.pdb -r B113 -o 1tupVAL113_only.pdb
1tupPRO111_only.pdb 1tupVAL113_only.pdb
- UCSF-Chimera has python interface which can be used to mutate residue.
- A python script (ChimeraMutateResidue.py) is made to mutate residue and write the mutated residue to a pdb file
- UCSF-Chimera package
- python module pychimera
pychimera ChimeraMutateResidue.py -f <PDB file> -r <RESIDUE number> -c <CHAIN name> -m <MUTANT (3-letter) residue> -o <OUTPUT file>
- integration of scripts into iCn3D web-framework
- Wang, Jiyao, et al. "iCn3D, a web-based 3D viewer for sharing 1D/2D/3D representations of biomolecular structures." Bioinformatics 36.1 (2020): 131-135.
- Xiang, Zhexin, and Barry Honig. "Extending the accuracy limits of prediction for side-chain conformations." Journal of molecular biology 311.2 (2001): 421-430.
Find the symmetry of a structure (assembly,chain,domain) and display it in iCn3D. We could convert the source code of SYMM to a web service.
- Read the papers: https://academic.oup.com/nar/article/42/W1/W296/2435489 and https://pubmed.ncbi.nlm.nih.gov/27747230/. The web server is at https://galaxyproject.org/use/symd/.
- The output of SYMD for the PDB 1KQ2 is at symd-1KQ2.txt. There are four parts.
- The Z1 score threshold (in parts 2 and 3 of the output) should be 10 (or 8). If all Z1 scores are less than 10, the structure has no symmetry.
- The symmetry from RCSB is not complete. It didn't show the local symmetry: https://structure.ncbi.nlm.nih.gov/icn3d/share.html?bGH1BfLsiGFhhTDn8.
- Based on the ouput above, determine how many symmetries in the structure.

- What are the symmetries, such as C6, D2, etc.
- Calculate the axis for each symmetry.
- Display the symmetry in iCn3D.
- You can test at https://www.ncbi.nlm.nih.gov/Structure/icn3d2/full.html?mmdbid=1kq2 by replacing 1kq2 with any pdb id.
- Click the menu "Analysis > Symmetry (SymD)". A popup window is shown. Click "Apply" to show the symmetry.
- Click "Ctrl + Shift + i" to see the "console" tab. The output from SymD is shown in console.
- You can select a subset of the structure to do this dynamic symmetry calculation as well.
- So far it only works for cyclic symmetry. We need to test with more examples and improve it.
About 15 structures were tested. 7 look good. Errors with other structures. Details here
- CE-Symm: https://www.sciencedirect.com/science/article/pii/S0022283614001557
- CE-Symm: https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1006842
Jiyao Wang (github: jiywang3)
Emily Tai
Top Madeji
Sridhar Acharya Malkaram (github: sridharacharya)
David Enoma (github: davidenoma)
Rachel Dunn (github: racheldunn)
Varun Kotipalli (github: varun-kotipalli)