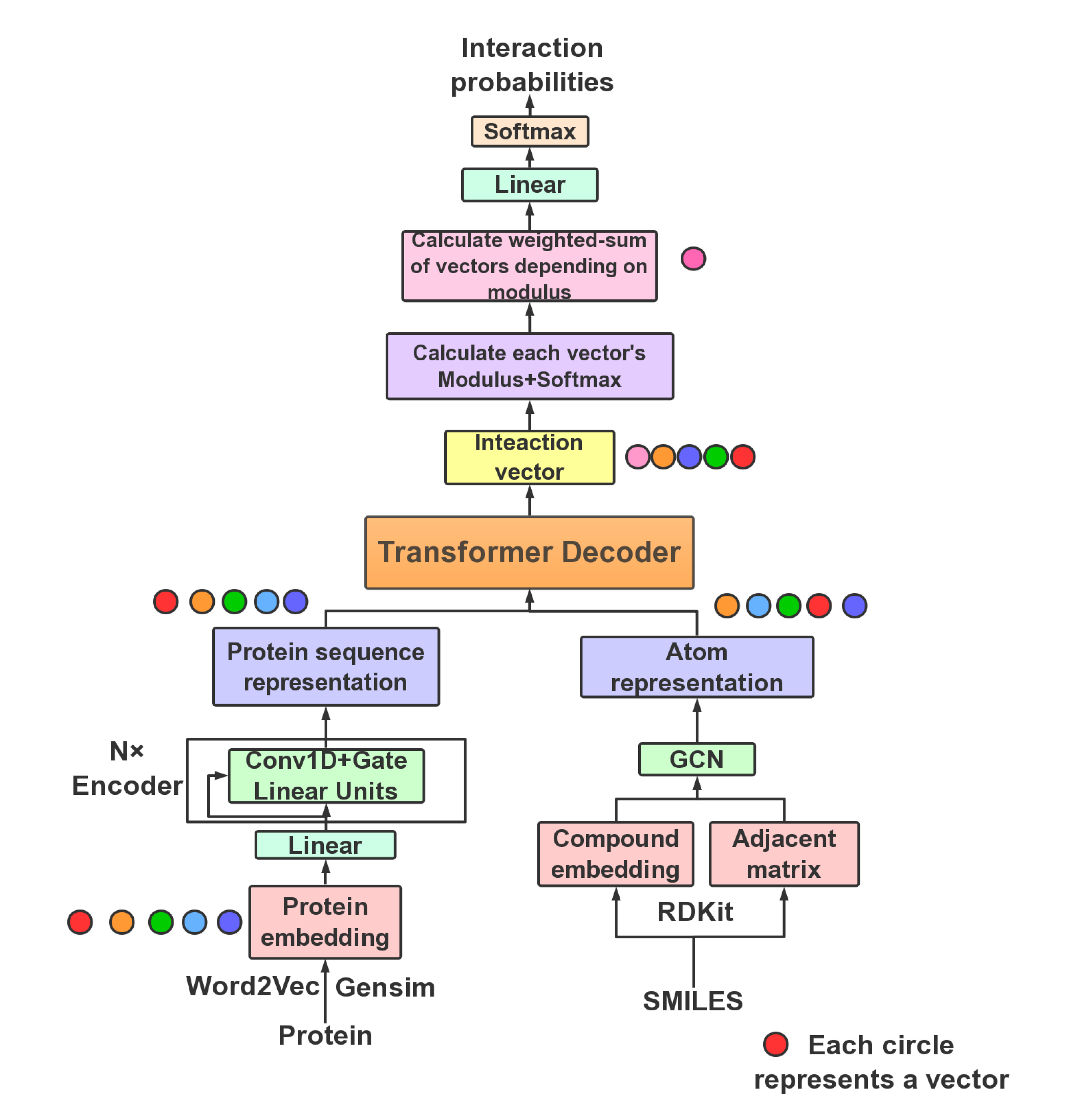
TransformerCPI: Improving compound–protein interaction prediction by sequence-based deep learning with self-attention mechanism and label reversal experiments
This repository contains the source code and the data.
TransformerCPI
Setup and dependencies
Dependencies:
- python 3.6
- pytorch >= 1.2.0
- numpy
- RDkit = 2019.03.3.0
- pandas
- Gensim >=3.4.0
Data sets
The data sets with train/test splits are provided as .7z file in a directory called 'data'.
The test set is created specially for label reversal experiments.
Using
1.mol_featurizer.py generates input for TransformerCPI model.
2.main.py trains TransformerCPI model.
Author
Lifan Chen (s18-chenlifan@simm.ac.cn)
Mingyue Zheng(myzheng@simm.ac.cn)
Citation
Lifan Chen, Xiaoqin Tan, Dingyan Wang, Feisheng Zhong, Xiaohong Liu, Tianbiao Yang, Xiaomin Luo, Kaixian Chen, Hualiang Jiang, Mingyue Zheng, TransformerCPI: Improving compound–protein interaction prediction by sequence-based deep learning with self-attention mechanism and label reversal experiments, Bioinformatics, , btaa524, https://doi.org/10.1093/bioinformatics/btaa524