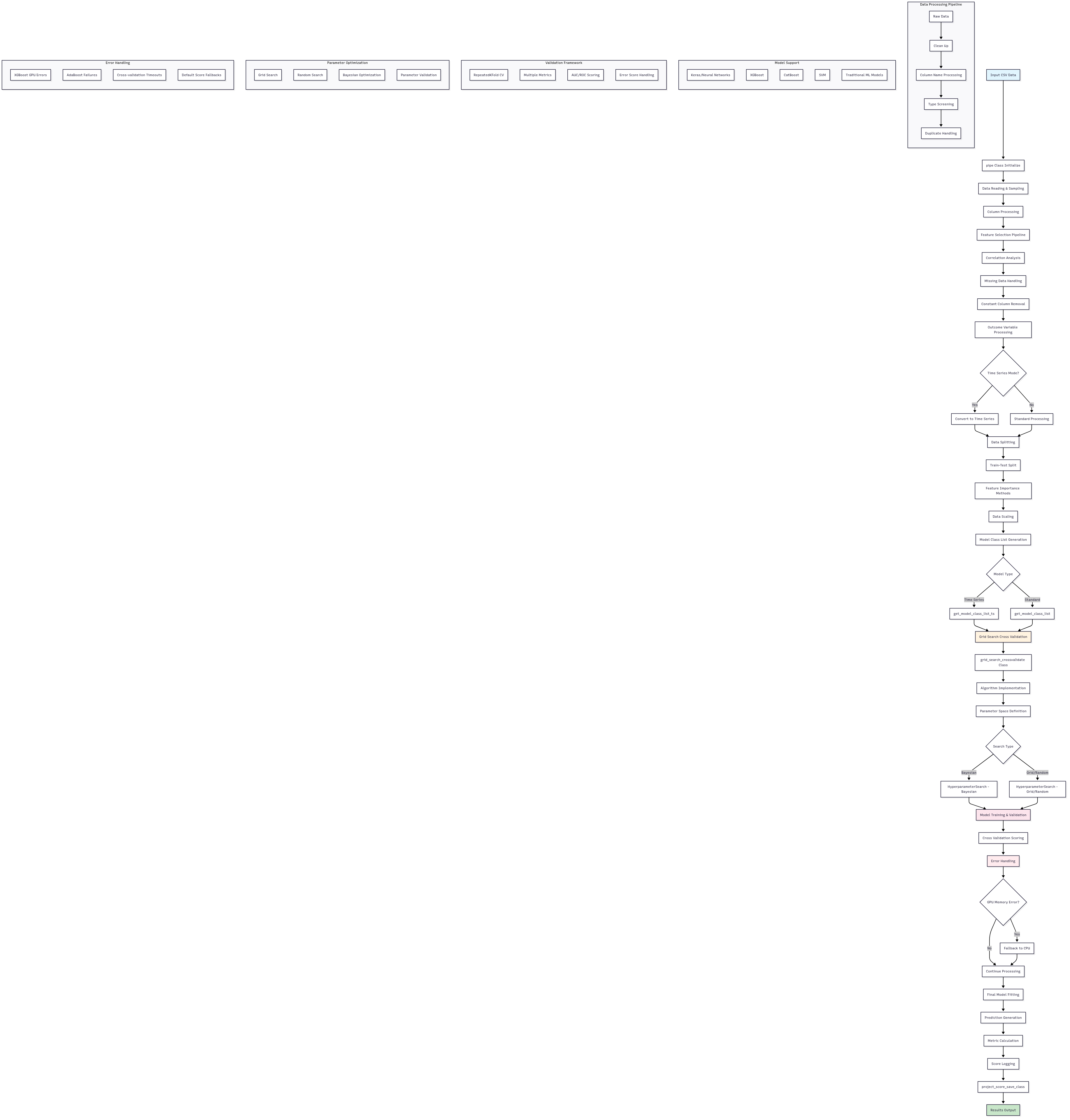
This repository contains Python code for binary classification using grid search and hyperparameter optimization techniques.
- ml_binary_classification_gridsearch_hyperOpt
- Overview
- Diagrams
- Features
- Getting Started
- Installation
- Usage
- Examples
- Project Structure
- Contributing
- License
- Appendix
- Acknowledgments
Binary classification is a common machine learning task where the goal is to categorize data into one of two classes. This repository provides a framework for performing binary classification using various machine learning algorithms and optimizing their hyperparameters through grid search and hyperparameter optimization techniques.
This framework is designed to be a comprehensive toolkit for binary classification experiments, offering a wide range of configurable options:
- Diverse Model Support: Includes a collection of standard classifiers (e.g., Logistic Regression, SVM, RandomForest, XGBoost, LightGBM, CatBoost) and specialized time-series models from the
aeonlibrary (e.g., HIVE-COTE v2, MUSE, OrdinalTDE). - Advanced Hyperparameter Tuning: Supports multiple search strategies:
- Grid Search: Exhaustively search a defined parameter grid.
- Random Search: Randomly sample from the parameter space.
- Bayesian Optimization: Intelligently search the parameter space using
scikit-optimize.
- Configurable Data Pipeline: A highly modular pipeline allows for fine-grained control over data processing steps:
- Feature Selection: Toggle groups of features (e.g., demographics, blood tests, annotations).
- Data Cleaning: Handle missing values, constant columns, and correlated features.
- Resampling: Address class imbalance with oversampling (RandomOverSampler) or undersampling (RandomUnderSampler).
- Scaling: Apply standard scaling to numeric features.
- Automated Results Analysis: Includes tools to automatically aggregate results from multiple runs and generate insightful plots, such as global parameter importance.
- Time-Series Capabilities: Specialized pipeline mode for handling time-series data, including conversion to the required 3D format for
aeonclassifiers.
Below are visual diagrams representing various components of the project. All .mmd source files are Mermaid diagrams, and the rendered versions are available in .svg or .png formats.
Designed for usage with a numeric data matrix for binary classification. Single or multiple outcome variables (One v rest). An example is provided. Time series is also implemented.
This project includes convenient installation scripts for Unix/Linux/macOS and Windows. These scripts will create a virtual environment, install all necessary dependencies, and register a Jupyter kernel for you.
-
Clone the repository:
git clone https://github.com/SamoraHunter/ml_binary_classification_gridsearch_hyperOpt.git cd ml_binary_classification_gridsearch_hyperOpt -
Run the installation script:
-
For a standard installation:
- On Unix/Linux/macOS:
chmod +x install.sh ./install.sh
- On Windows:
install.bat
This will create a virtual environment named
ml_grid_env. - On Unix/Linux/macOS:
-
For a time-series installation (includes all standard dependencies):
- On Unix/Linux/macOS:
chmod +x install.sh ./install.sh ts
- On Windows:
install.bat ts
This will create a virtual environment named
ml_grid_ts_env. - On Unix/Linux/macOS:
-
After installation, activate the virtual environment to run your code or notebooks.
-
To activate the standard environment:
- On Unix/Linux/macOS:
source ml_grid_env/bin/activate - On Windows:
.\ml_grid_env\Scripts\activate
- On Unix/Linux/macOS:
-
To activate the time-series environment:
- On Unix/Linux/macOS:
source ml_grid_ts_env/bin/activate - On Windows:
.\ml_grid_ts_env\Scripts\activate
- On Unix/Linux/macOS:
The notebooks/ directory contains examples for different use cases:
unit_test_synthetic.ipynb: The main entry point for running experiments. It demonstrates how to generate synthetic data, test the data pipeline, and run a full Hyperopt search. Start here to understand the end-to-end workflow.01_hyperopt_grid.ipynb: A focused example of running a full hyperparameter search usingHyperoptbased on theconfig_hyperopt.ymlfile.02_single_run.ipynb: A script for executing a single run with a specific set of parameters defined inconfig_single_run.yml. Useful for debugging or testing one configuration.
The main entry point for running experiments is a script or notebook that loads the configuration and iterates through the parameter space defined in config.yml.
-
Configure your experiment in
config.yml:- Set the data path, models, and parameter space.
-
Run the experiment:
- The following script demonstrates how to execute a full grid search based on your
config.yml.
- The following script demonstrates how to execute a full grid search based on your
from pathlib import Path
from ml_grid.pipeline.data import pipe
from ml_grid.util.param_space import parameter_space
from ml_grid.util.create_experiment_directory import create_experiment_directory
from ml_grid.util.config_parser import load_config
# Load configuration from config.yml
config = load_config()
# Set project root
project_root = Path().resolve().parent
# Create a unique directory for this experiment run
experiments_base_dir = project_root / config['experiment']['experiments_base_dir']
experiment_dir = create_experiment_directory(
base_dir=experiments_base_dir,
additional_naming=config['experiment']['additional_naming']
)
# Generate the parameter space from the config file
param_space_df = parameter_space(config['param_space']).get_parameter_space()
# Iterate through each parameter combination and run the pipeline
for i, row in param_space_df.iterrows():
local_param_dict = row.to_dict()
print(f"Running experiment {i+1}/{len(param_space_df)} with params: {local_param_dict}")
pipe(
config=config,
local_param_dict=local_param_dict,
base_project_dir=project_root,
experiment_dir=experiment_dir,
param_space_index=i
)If you are using Jupyter, you can also select the kernel created during installation (e.g., Python (ml_grid_env)) directly from the Jupyter interface.
See [ml_grid/tests/unit_test_synthetic.ipynb]
The latest documentation is hosted online and can be viewed here.
This project uses Sphinx for documentation. The documentation includes usage guides and an auto-generated API reference.
To build the documentation locally:
-
Install the documentation dependencies (make sure your virtual environment is activated):
pip install -e .[docs]
-
Build the HTML documentation:
sphinx-build -b html docs/source docs/build
-
Open
docs/build/index.htmlin your web browser to view the documentation.
The repository is organized to separate concerns, making it easier to navigate and extend.
.
├── assets/ # Mermaid diagrams and other assets
├── docs/ # Sphinx documentation source and build files
├── ml_grid/ # Main source code for the library
│ ├── model_classes/ # Standard classifier wrappers
│ ├── model_classes_time_series/ # Time-series classifier wrappers
│ ├── pipeline/ # Core data processing and pipeline logic
│ ├── results_processing/ # Tools for aggregating and plotting results
│ └── util/ # Utility functions and global parameters
├── tests/ # Unit and integration tests
├── install.sh # Installation script for Unix/Linux
└── install.bat # Installation script for Windows
If you would like to contribute to this project, please follow these steps:
Fork the repository on GitHub. Create a new branch for your feature or bug fix. Make your changes and commit them with descriptive commit messages. Push your changes to your fork. Create a pull request to the main repository's master branch.
This project is licensed under the MIT License - see the LICENSE file for details.
scikit-learn hyperopt