This repo contains the official implementation of the paper Any-Property-Conditional Molecule Generation with Self-Criticism using Spanning Trees with our method called STGG+, an improvement over the original STGG method by Sungsoo Ahn et al (2022). See also our blog post for more information.
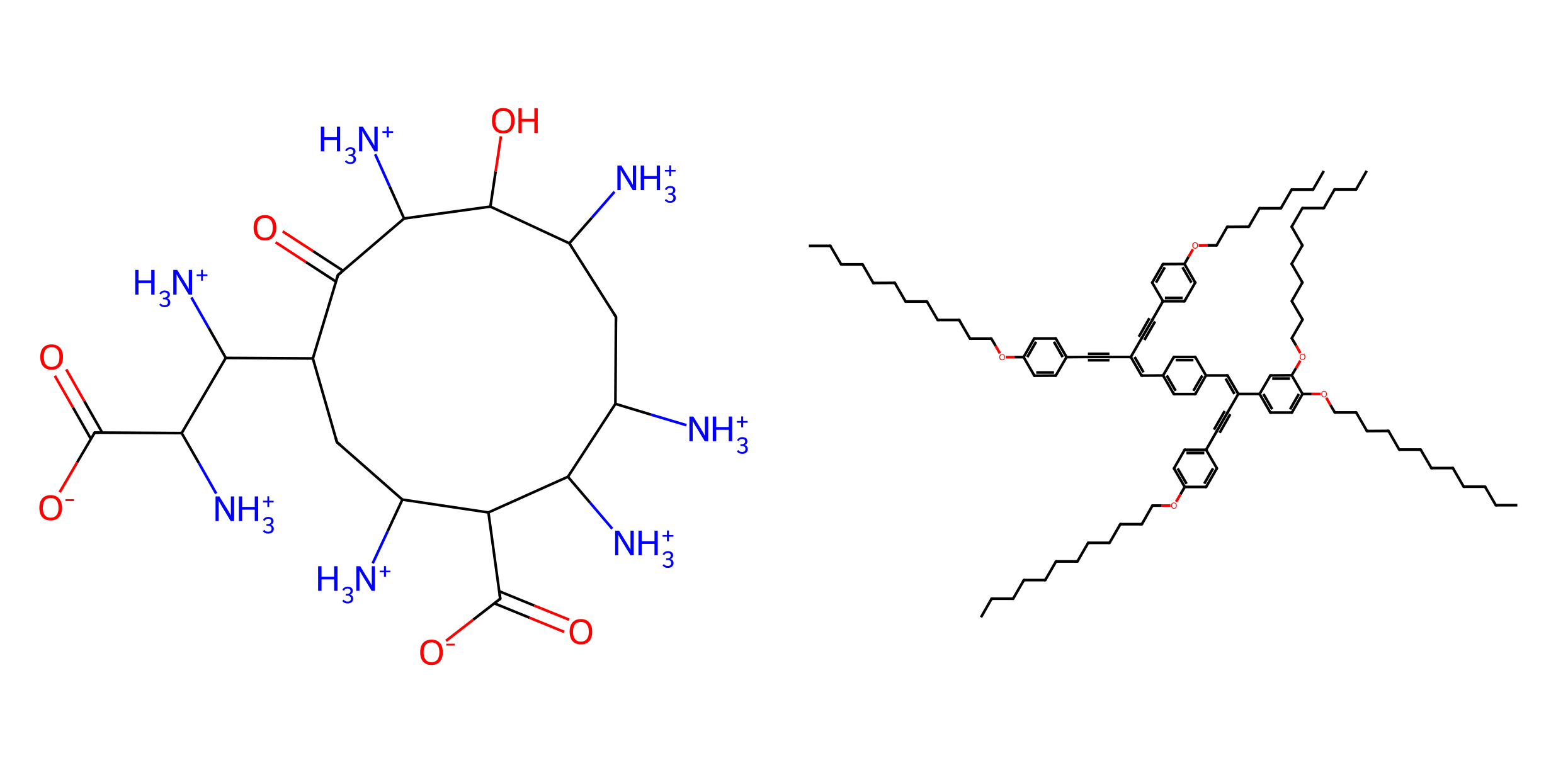 Molecules generated by our STGG+ model. Left: conditioned on logP=-13.6292; Right: conditioned on logP=28.6915. logP is related to water solubility, so the left one is very hard to dissolve in water, while the right one is easy to dissolve.
Molecules generated by our STGG+ model. Left: conditioned on logP=-13.6292; Right: conditioned on logP=28.6915. logP is related to water solubility, so the left one is very hard to dissolve in water, while the right one is easy to dissolve.
You need to get a (free) neptune account and modify the YOUR_API_KEY and YOUR_PROJECT_KEY for neptune initialization in train.py.
You need to set the directory location of your model checkpoints by modifying the default value for --save_checkpoint_dir in train.py.
You must install all the requirements below and build the vocabulary and valencies for each dataset.
## Make env from scratch
module load python/3.10
module load cuda/11.8
python -m venv your_dir/molecules_autoregressive
source your_dir/molecules_autoregressive/bin/activate
pip install --upgrade pip setuptools wheel
pip install --upgrade --pre torch=2.3.0 torchvision torchaudio --index-url https://download.pytorch.org/whl/cu118
pip install lightning neptune
pip install torch-geometric
pip install pyg_lib torch_scatter torch_sparse torch_cluster torch_spline_conv -f https://data.pyg.org/whl/torch-2.3.0+cu118.html
pip install cython molsets rdkit pomegranate==0.14.8 pyyaml scikit-learn pandas numpy networkx
pip install fcd_torch
git clone https://github.com/AlexiaJM/moses_fixed # fix two annoying bugs in MOSES, which is not updated anymore
cd moses_fixed
python setup.py install
## Build vocabulary
python data_preprocessing.py --dataset_name qm9 --MAX_LEN 150 --force_vocab_redo
python data_preprocessing.py --dataset_name zinc --MAX_LEN 250 --force_vocab_redo
python data_preprocessing.py --dataset_name chromophore --MAX_LEN 750 --force_vocab_redo # max-length = 511
# Dit paper
CUDA_VISIBLE_DEVICES=0 python data_preprocessing_dit.py --dataset_name bace --MAX_LEN 300 --force_vocab_redo --num_workers your_number_of_cpus # max_len=144
CUDA_VISIBLE_DEVICES=0 python data_preprocessing_dit.py --dataset_name bbbp --MAX_LEN 300 --force_vocab_redo --num_workers your_number_of_cpus # max_len=180
CUDA_VISIBLE_DEVICES=0 python data_preprocessing_dit.py --dataset_name hiv --MAX_LEN 300 --force_vocab_redo --num_workers your_number_of_cpus # max_len=144
# reward maximization (reproducing Multi-Objective GFlowNets)
CUDA_VISIBLE_DEVICES=0 python data_preprocessing_gflownet.py --dataset_name qm9 --MAX_LEN 150 # max-length = 48
To reproduce the examples from the paper, you can run examples from experiments/exps.sh.
Its relatively straightforward, but there are lots of options to play with. I recommend to always use as options: --lambda_predict_prop 1.0 --randomize_order --start_random --scaling_type std --special_init --swiglu --gpt --no_bias --rmsnorm --rotary (for STGG+). At test time, you can tune the guidance (which I did not tune and left to 1.5 as default).
Here is an example below for the Chromophore OOD training:
cd /src
# Training
CUDA_VISIBLE_DEVICES=0 python train.py --dataset_name chromophore --num_layers 3 --tag exp_chromophore50epoch --bf16 \
--check_sample_every_n_epoch 999 --dropout 0.0 --warmup_steps 100 --lr_decay 0.1 --beta2 0.95 --weight_decay 0.1 --lambda_predict_prop 1.0 \
--batch_size 128 --lr 2.5e-4 --max_epochs 1000 --n_gpu 1 --randomize_order --start_random --scaling_type std --special_init --nhead 16 --swiglu --expand_scale 2.0 \
--max_len 600 --gpt --no_bias --rmsnorm --rotary --log_every_n_steps 24
# Testing respectively with guidance=1.5 for k=1, k=100, random-guidance for k=1, k=100.
CUDA_VISIBLE_DEVICES=0 python train.py --dataset_name chromophore --num_layers 3 --tag exp_chromophore1000epoch --bf16 \
--check_sample_every_n_epoch 999 --dropout 0.0 --warmup_steps 100 --lr_decay 0.1 --beta2 0.95 --weight_decay 0.1 --lambda_predict_prop 1.0 \
--batch_size 128 --lr 2.5e-4 --max_epochs 1000 --n_gpu 1 --randomize_order --start_random --scaling_type std --special_init --nhead 16 --swiglu --expand_scale 2.0 \
--max_len 600 --gpt --no_bias --rmsnorm --rotary \
--guidance 1.5 --guidance_ood 1.5 --only_ood --no_test_step --not_allow_empty_bond \
--test --ood_values 1538 -531 28.6915 -13.6292 1.2355 -0.5406 --sample_batch_size 100 --num_samples_ood 100 --best_out_of_k 1
CUDA_VISIBLE_DEVICES=0 python train.py --dataset_name chromophore --num_layers 3 --tag exp_chromophore50epoch --bf16 \
--check_sample_every_n_epoch 999 --dropout 0.0 --warmup_steps 100 --lr_decay 0.1 --beta2 0.95 --weight_decay 0.1 --lambda_predict_prop 1.0 \
--batch_size 128 --lr 2.5e-4 --max_epochs 1000 --n_gpu 1 --randomize_order --start_random --scaling_type std --special_init --nhead 16 --swiglu --expand_scale 2.0 \
--max_len 600 --gpt --no_bias --rmsnorm --rotary \
--guidance 1.5 --guidance_ood 1.5 --only_ood --no_test_step --not_allow_empty_bond \
--test --ood_values 1538 -531 28.6915 -13.6292 1.2355 -0.5406 --sample_batch_size 100 --num_samples_ood 100 --best_out_of_k 100
CUDA_VISIBLE_DEVICES=0 python train.py --dataset_name chromophore --num_layers 3 --tag exp_chromophore50epoch --bf16 \
--check_sample_every_n_epoch 999 --dropout 0.0 --warmup_steps 100 --lr_decay 0.1 --beta2 0.95 --weight_decay 0.1 --lambda_predict_prop 1.0 \
--batch_size 128 --lr 2.5e-4 --max_epochs 1000 --n_gpu 1 --randomize_order --start_random --scaling_type std --special_init --nhead 16 --swiglu --expand_scale 2.0 \
--max_len 600 --gpt --no_bias --rmsnorm --rotary \
--guidance 1.5 --guidance_ood 1.5 --guidance_rand --only_ood --no_test_step --not_allow_empty_bond \
--test --ood_values 1538 -531 28.6915 -13.6292 1.2355 -0.5406 --sample_batch_size 100 --num_samples_ood 100 --best_out_of_k 1
CUDA_VISIBLE_DEVICES=0 python train.py --dataset_name chromophore --num_layers 3 --tag exp_chromophore50epoch --bf16 \
--check_sample_every_n_epoch 999 --dropout 0.0 --warmup_steps 100 --lr_decay 0.1 --beta2 0.95 --weight_decay 0.1 --lambda_predict_prop 1.0 \
--batch_size 128 --lr 2.5e-4 --max_epochs 1000 --n_gpu 1 --randomize_order --start_random --scaling_type std --special_init --nhead 16 --swiglu --expand_scale 2.0 \
--max_len 600 --gpt --no_bias --rmsnorm --rotary \
--guidance 1.5 --guidance_ood 1.5 --guidance_rand --only_ood --no_test_step --not_allow_empty_bond \
--test --ood_values 1538 -531 28.6915 -13.6292 1.2355 -0.5406 --sample_batch_size 100 --num_samples_ood 100 --best_out_of_k 100
The useful arguments that you can provide to train.py are as follows (yes, there are a lot!):
# Important base arguments
--save_checkpoint_dir=my_path # save directory for checkpoints (Important to change to your own directory)
--tag=my_exp_zinc_50_epoch # name you give to your model/experiment
--n_gpu=1 # number of GPUs
--cpu # use CPUs instead of GPUs
--seed=666 # seed
--dataset_name=zinc # zinc, qm9, moses, chromophore, hiv, bbbp, bace
--num_workers=6 # number of CPU workers per GPU (24 workers with 4 GPUs or 6 workers with 1 GPU = set it to 6)
--max_len=250 # Max sentence length, please make sure that its high enough based on the data_preprocessing.py showing the dataset approximate max-length. Add around 25% more to handle out-of-distribution cases. The max_len cannot be changed from training to sampling time, so make sure to make it big enough. But keep in mind, bigger max_len makes things slower.
# Things to always leave on
--bf16 # use BF16 instead of FP32
--lambda_predict_prop=1.0 # property-predictor with loss weighting of 1.0
--randomize_order # randomize the order of the graph nodes
--start_random # randomize the start node of the graph (otherwise it start from one node with the least amount of neighboors)
--scaling_type=std # (none, std) std standardize the properties (z-score)
--gpt # Uses a Modern Transformer
--special_init # use the GPT-3 proj_weight init
--swiglu # use SwiGLU
--no_bias # remove the bias terms
--rmsnorm # use RMSNorm instead of LayerNorm
--rotary # use Rotary embedding instead of relative positional embedding
# Options for fine-tuning
--finetune_dataset_name='' # when not empty, the dataset is used for fine-tuning
--finetune_scaler_vocab # If True, uses the fine-tuning dataset vocab instead of the pre-training dataset vocab for z-score standardization
--load_checkpoint_path=my_path # load a specific checkpoint; use it to fine-tune
# Model hyperparameters
--num_layers=3 # number of layers
--emb_size=1024 # embedding size
--nhead=16 # number of attention heads
--expand_scale=2.0 # expand factor for the MLP in the Modern Transformer
--dropout=0.0 # dropout
# Optimizer hyperparameters
--max_epochs # number of epochs
--batch_size=512 # batch-size for training
--lr=1e-3 # learning rate, set propertionaly to batch-size
--warmup_steps=200 # a few warmup steps is supposedly good
--lr_decay=0.1 # cosine learning decay from lr to lr_decay*lr (use 0.1 for LLMs)
--beta1=0.9 # AdamW beta1
--beta2=0.95 # AdamW beta1=0.95 for LLMs, otherwise 0.999 is standard
--weight_decay=0.1 # 0.1 for LLMs
--gradient_clip_val=1.0 # gradient clipping
# Sampling
--test # do sampling instead of training
--sample_batch_size=1250 # batch-size for sampling
--num_samples=10000 # number of samples for in-distribution metrics
--num_samples_ood=2000 # number of samples for out-of-distribution metrics
# Sampling tunable knobs
--best_out_of_k=1 # If >1, we sample k molecules and choose the best-out-of-k based on the unconditional model property predictions (when using --lambda_predict_prop 1.0)
--temperature=1.0 # temperature for in-distribution
--temperature_ood=1.0" # temperature for out-of-distribution
--guidance=1.0 # classifier-free-guidance value (1.0 means no guidance)
--guidance_ood=1.0 # classifier-free-guidance value (1.0 means no guidance)
--guidance_rand # If True, randomly chooses a guidance between [0.5, 2]
--top_k=0 # If > 0, we only select from the top-k tokens
# Sampling other options
--not_allow_empty_bond # use to disable empty bonds; when there are compounds in the data, we need the empty-bond token during training, but this option can be used to prevent creating compounds at sampling time.
--no_ood # Do not do out-of-distribution sampling (only do in-distribution)
--only_ood # Only do out-of-distribution sampling (not in-distribution)
--no_test_step # Ignore the test set, useful to speed up things when doing --only_ood
--ood_values 580 84 8.194 -3.281 1.2861 0.1778 # Manually set out-of-distribution values (otherwise, they are automatically calculated from the training dataset as +- 4 standard-deviation): needs to be [max,min] for all properties, so should be 6 values if you have 3 properties; for Zinc based on https://arxiv.org/pdf/2208.10718, the values are 580, 84, 8.194, -3.281, 1.2861, 0.1778.
# Less important base arguments
--compile # compile for improve performance, but at the current time, it does not work, torch.compile is so buggy
--log_every_n_steps # control how often to log on Neptune
--n_correct=20 # max_len=250 means that at len = 240 - n_correct - number_of_currently_opened_branches, we force the spanning-tree to close all its branches ASAP to prevent an incomplete spanning-tree (choose it big enough to prevent incomplete samples, I found that 20 was always good enough)
--check_sample_every_n_epoch=10 # how often (in epochs) to run the sampling metrics (default every 10 epochs, we check the metrics). Honestly, you can leave it at 999999 because the metrics are generally best the longer you train.
If you find the code useful, please consider citing our STGG+ paper:
@misc{jolicoeurmartineau2024anyproperty,
title={Any-Property-Conditional Molecule Generation with Self-Criticism using Spanning Trees},
author={Alexia Jolicoeur-Martineau and Aristide Baratin and Kisoo Kwon and Boris Knyazev and Yan Zhang},
year={2024},
eprint={2407.09357},
archivePrefix={arXiv},
primaryClass={cs.LG}
}and the original STGG paper:
@inproceedings{ahn2022spanning,
title={Spanning Tree-based Graph Generation for Molecules},
author={Sungsoo Ahn and Binghong Chen and Tianzhe Wang and Le Song},
booktitle={International Conference on Learning Representations},
year={2022},
url={https://openreview.net/forum?id=w60btE_8T2m}
}Note that this code is based on the original STGG code, which can be found in the Supplementary Material section of https://openreview.net/forum?id=w60btE_8T2m.