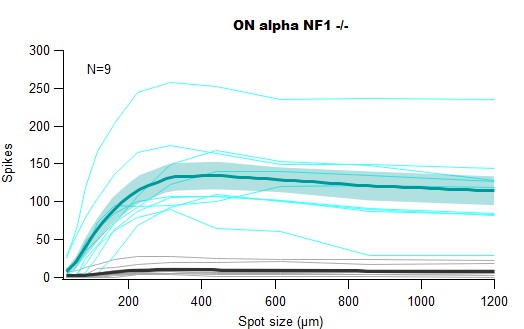
For these experiments, we recorded from the three Alphas ganglion cell types in loose-seal cell attached configuration while presenting light stimuli from darkness of different diamaters (30 to 1200 um). We measured:
- Baseline firing rate before the light stimuli
- Peak firing rates (Hz)
- Peak Spot Size: the size of the light stimuli which elicited the most spikes.
- Peak spike count: maximum number of spikes elicited by the light stimuli.
Example figure:
-
These are created directly from the
NF1.h5file located in Data_final. -
It has the following hierarchy: animal_type (NF1_control, NF1_het, NF1_homo)-> celltype -> cellname.
-
For each cell, on the Y axis we plotted
spikes_stim_meanfor the ON response (cyan) andspikes_tail_meanfor the OFF response (gray) and then computed the mean (darker lines). The SEM (shaded areas) are also part of the HDF5 filespikes_stim_semandspikes_tail_semfor the ON and OFF response, respectiviel.
Example figure:
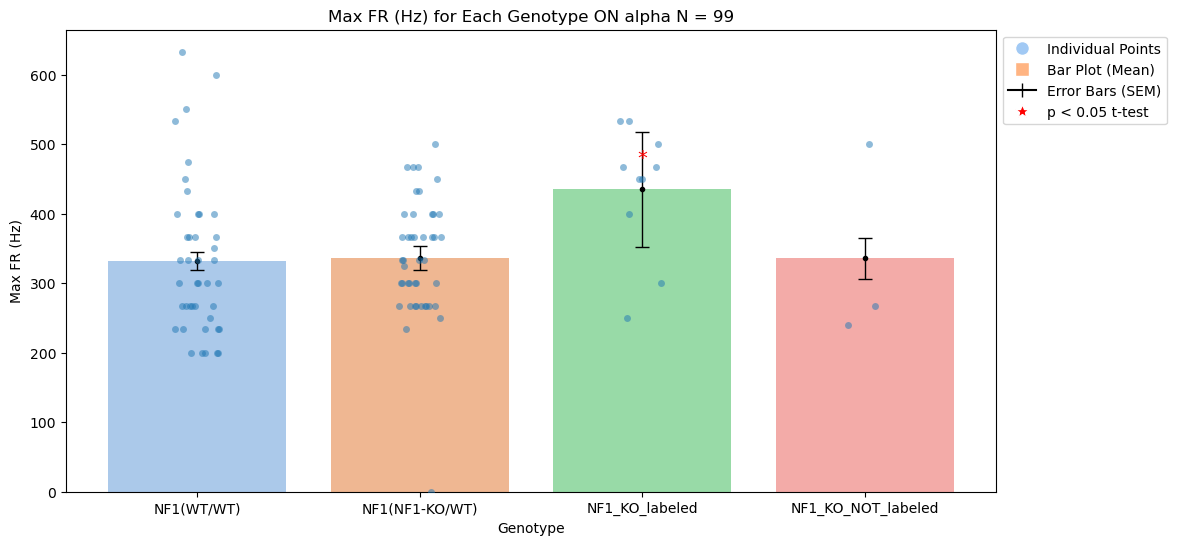
In the result_tables, I have separated the two cohort analysis. In the result_tables/First_cohort directory there are .csv files for every celltype which include the relevant genotype,sex, eye side,quadrant and cell_type information and it's responses. Attached is also the Jupyer Notebook NF1_Analysis used to create plot the individual points, mean and run t-tests correcting for multiple comparisons.
Example figure:
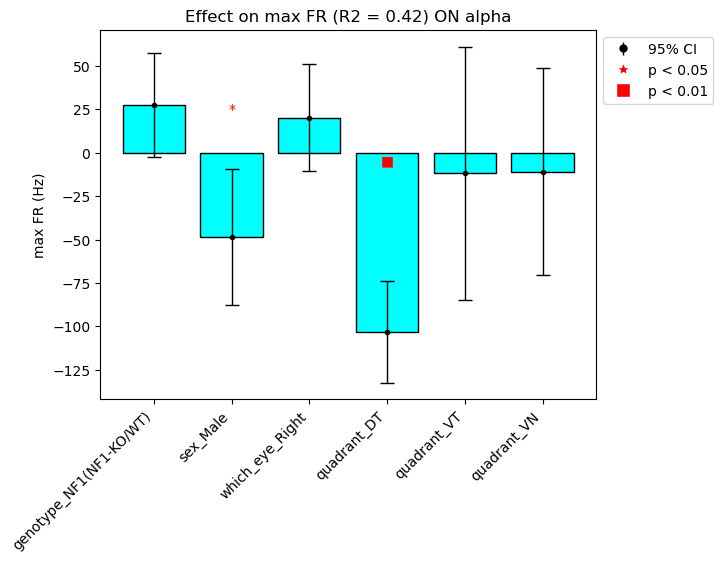
We ran linear mixed effects models using the fitlme function in Matlab and exported it results as CSVs result_tables/First_cohort for each celltype (separate directories). Also attached isthe Jupyter Notebook LME_results I used to create the plots.