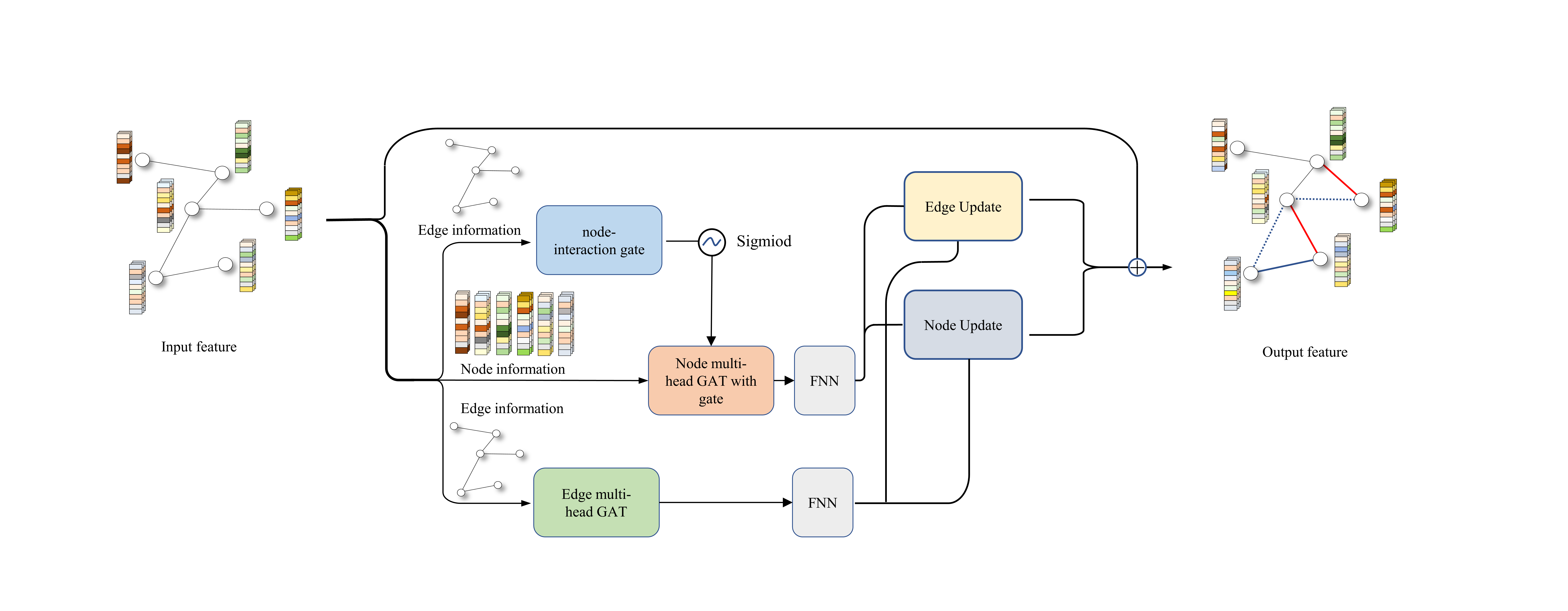
A highly sensitive model based on graph neural networks for enzyme key catalytic residues prediction
A new enzyme catalytic sites prediction model use new designed Adaptive Edge-Gated Graph Attention Neural Network(AEGAN) to process both sequence and structure features of proteins at different levels was developed here.
torch==1.10.0
tqdm==4.62.3
numpy==1.23.2
pytorch-lightning==1.7.2
biotite==0.34.1
pandas==1.3.4
scikit-learn==1.1.1
psiblast==2.12.0+
- First, install BlAST using:
conda install -c bioconda blast - Download protein database for alignment, using:
wget ftp://ftp.uniprot.org/pub/databases/uniprot/uniref/uniref90/uniref90.fasta.gz - compile the protein database as follows:
gzip -d uniref90.fasta.gz makeblastdb -in uniref90.fasta -parse_seqids -hash_index -dbtype prot - and then,
cd AEGAN/code,you can create datasets, using
DataProcess.py -s uni14230 -d your-database-path
there are some parameters you can choose:-s:specifies which dataset to createuni14230train datasetuni3175test datasetEF_foldbenchmark datasetEF_familybenchmark datasetEF_superfamilybenchmark datasetHA_superfamilybenchmark datasetNNbenchmark datasetPCbenchmark dataset
-t:(optional) specify where the dataset will be saved-d:database path used for BLAST alignment
cd AEGAN/code, and use train.py to train a model:python train.py -sm model-saved-path
there are some parameters you can choose:--trainset:The path of the dataset of the training model (default:../database/trainset)--batchsize:batchsize (default:200)-lr:learning rate (default:1e-2)-ly:layers of AEGAN module (default:24)--epoch:Traing epochs (default:1000)-sm:Path of saving trained model--accelerator: Uesd device to train model (default:gpu)cpu:use cpu to train modelgpu:use gpu to train model
cd AEGAN/code, and use validate.py to test model's performance on datasets:
python validate.py --model trained-model-path- if you haven't trained a model yet, you can use the model we've trained to test it:
python validate.py --model trained_full - and you can also get the results of ablation experiment using the trained model:
python validate.py --model EP --lack PSSM
there are also some parameters you can choose:--model:specifies the path of the model to test. it can be a model file or folder contains model files.trained_full:trained full features modelEP:trained model which features exclude PSSMEAT:trained model which features exclude AtchleyEAM:trained model which features exclude AtomModel
--testset:Test dataset (default:uni3175)uni3175EF_foldEF_familyEF_superfamilyHA_superfamilyNNPC
--lack:specify which feature is missingPSSMAtchleyAtomModel
--accelerator: Uesd device to train model (default:gpu)cpu:use cpu to train modelgpu:use gpu to train model
--batchsize:batchsize (default:100)