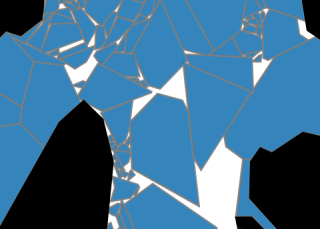
KSPoly utilizes Gaussian kernels to create a smooth representation of scalar, vector, or tensor fields defined on a set of polygons within a 2D region. The input frame consist of a collection of polygons in the plane, each paired with its corresponding field quantities, which could come from output generated by a DEM model or observational data. By triangulating the polygons, KSPoly seamlessly integrates the fields using a Gaussian kernel with the scaling parameter
You can download and install KSPoly from github repository. The KSPoly module uses numba, numpy, scikit-learn and triangle libraries.
pip install git+https://github.com/SeaIce-Math/KSPoly.gitAlternative approach:
git clone https://github.com/SeaIce-Math/KSPoly.git
cd KSPoly
pip install .To use KSPoly, you need to use the parameters of the domain and the kernel function to construct a scene object. Then, for each frame, provide a list of L polygons (encoded by X and Y coordinates of their vertices) with associated field quantities of your choice (any tensor of the shape (L,...)). Finally, the smoothit() function outputs the smooth representation of the fields in the 2D domain.
import kspoly as ks
import numpy as np
from matplotlib import pyplot as plt
import matplotlib.cm as cm
# Construct the scene for polygon fields
sc = ks.scene(sigma = 0.4, mesh_size=[60,60], xlim = [-8,8], ylim = [-10,10])
# define a list of polygons
poly1 = np.array([[0,2,1],[0,0,2]])
poly2 = np.array([[5,6,6,4],[1,1,3,3]])
# specify the velocity for each polygon
v1 = np.array([-1,1])
v2 = np.array([1,0])
# specify the stress tensor for each polygon
st1 = np.array([[1,2],[2,1]])
st2 = np.array([[1,-1],[0,1]])
for i in range(3):
# move polygons based on time steps
poly1 = poly1 + 0.5*i*v1.reshape(-1,1)
poly2 = poly2 + 0.5*i*v2.reshape(-1,1)
# Create a new frame from the set of polygons with their corresponding velocity.
frame = ks.frame_data([poly1, poly2], velocities=[v1, v2])
# Add stress data
frame.set_field([st1, st2], name='stress')
# Add mass desnity associated to each polygon
frame.set_field([2, 3], name='mass')
# Perform the smoothing and construct the smooth representation
sm = sc.smoothit(frame)
sm_vel = sm['velocity']
sm_mass = sm['mass']
sm_stress = sm['stress']
# Make plots for smooth representation of mass and velocity fields
mass = np.squeeze(sm_mass)
v_x = np.squeeze(sm_vel[:,:,0])
v_y = np.squeeze(sm_vel[:,:,1])
fig, axs = plt.subplots(nrows=1, ncols=3, figsize=(18, 6), sharey=True, sharex =True, dpi=100)
axs[0].set_title("smooth mass density", fontsize=18)
h0 = axs[0].pcolormesh(sc.frame_xx, sc.frame_yy, mass, cmap=cm.Blues , vmin=0.0, vmax=1.0, alpha=1.0)
cbar = plt.colorbar(h0,ax=axs[0], ticks=[0, 1])
axs[1].set_title("smooth x component of velocity", fontsize=18)
h1 = axs[1].pcolormesh(sc.frame_xx, sc.frame_yy, v_x, cmap=cm.RdBu , vmin=-2.0, vmax=2.0, alpha=1.0)
cbar = plt.colorbar(h1,ax=axs[1], ticks=[-2, 2])
axs[2].set_title("smooth y component of velocity", fontsize=18)
h2 = axs[2].pcolormesh(sc.frame_xx, sc.frame_yy, v_y, cmap=cm.RdBu , vmin=-2.0, vmax=2.0, alpha=1.0)
cbar = plt.colorbar(h2,ax=axs[2], ticks=[-2, 2])
plt.show()