Protein Graph Library
This package provides functionality for producing a number of types of graph-based representations of proteins. We provide compatibility with standard formats, as well as graph objects designed for ease of use with popular deep learning libraries.
- Protein Graph Visualisation!
- RNA Graph Construction from Dotbracket notation
from graphein.construct_graphs import ProteinGraph
# Initialise ProteinGraph class
pg = ProteinGraph(granularity='CA', insertions=False, keep_hets=True,
node_featuriser='meiler', get_contacts_path='/Users/arianjamasb/github/getcontacts',
pdb_dir='examples/pdbs/',
contacts_dir='examples/contacts/',
exclude_waters=True, covalent_bonds=False, include_ss=True)
# Create residue-level graphs. Chain selection is either 'all' or a list e.g. ['A', 'B', 'D'] specifying the polypeptide chains to capture
# DGLGraph From PDB Accession Number
graph = pg.dgl_graph_from_pdb_code('3eiy', chain_selection='all')
# DGLGraph From PDB file
graph = pg.dgl_graph_from_pdb_file(file_path='examples/pdbs/pdb3eiy.pdb', contact_file='examples/contacts/3eiy_contacts.tsv', chain_selection='all')
# Create atom-level graphs
graph = pg._make_atom_graph(pdb_code='3eiy', graph_type='bigraph')from graphein.construct_meshes import ProteinMesh
# Initialise ProteinMesh class
pm = ProteinMesh()
# Pytorch3D Mesh Object from PDB Code
verts, faces, aux = pm.create_mesh(pdb_code='3eiy', out_dir='examples/meshes/')
# Pytorch3D Mesh Object from PDB File
verts, faces, aux = pm.create_mesh(pdb_file='examples/pdbs/pdb3eiy.pdb')from graphein.construct_graphs import RNAGraph
# Initialise RNAGraph Constructor
rg = RNAGraph()
# Build the graph from a dotbracket & optional sequence
rna = rg.dgl_graph_from_dotbracket('..(((((..(((...)))..)))))...', sequence='UUGGAGUACACAACCUGUACACUCUUUC')Graphs can be constructed according to walks through the graph in the figure below.
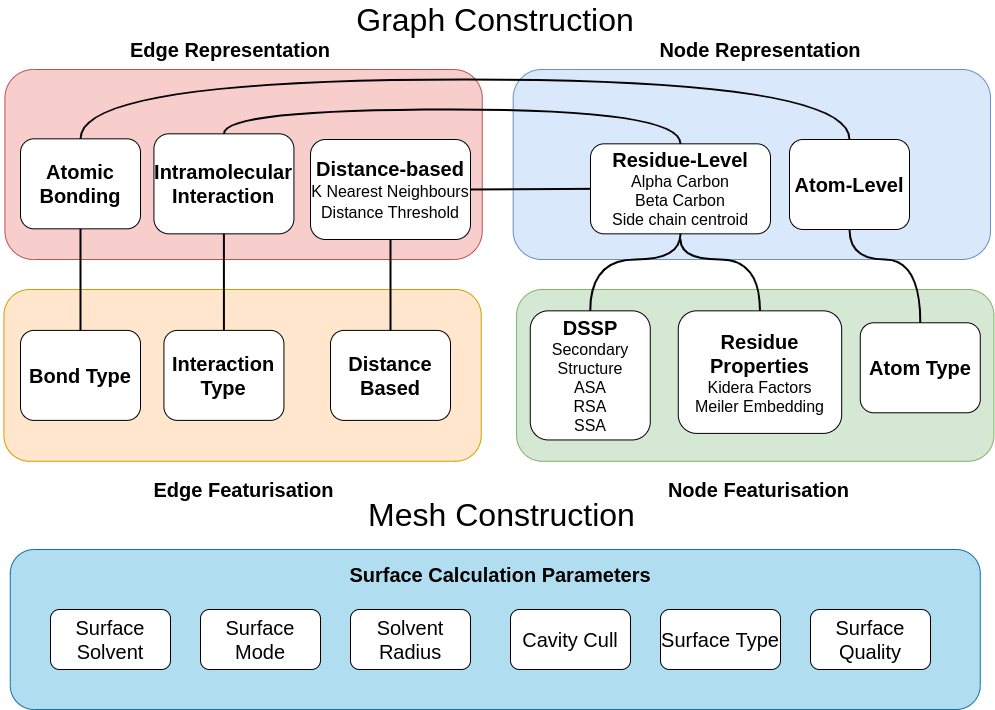
granularity: {'CA', 'CB', 'atom'} - specifies node-level granularity of graph
insertions: bool - keep atoms with multiple insertion positions
keep_hets: bool - keep hetatoms
node_featuriser: {'meiler', 'kidera'} low-dimensional embeddings of AA physico-chemical properties
pdb_dir: path to pdb files
contacts_dir: path to contact files generated by get_contacts
get_contacts_path: path to GetContacts installation
exclude_waters: bool - retain structural waters
covalent_bonds: bool - maintain covalent bond edges or just use intramolecular interactions
include_ss: bool - calculate protein SS and surface features using DSSP and assign them as node features
-
Create env:
conda create --name graphein python=3.7 conda activate graphein
-
Install GetContacts
# Install get_contact_ticc.py dependencies $ conda install scipy numpy scikit-learn matplotlib pandas cython seaborn $ pip install ticc==0.1.4 # Install vmd-python dependencies $ conda install netcdf4 numpy pandas seaborn expat tk=8.5 # Alternatively use pip $ brew install netcdf pyqt # Assumes https://brew.sh/ is installed # Install vmd-python library $ conda install -c conda-forge vmd-python # Set up getcontacts library $ git clone https://github.com/getcontacts/getcontacts.git $ echo "export PATH=`pwd`/getcontacts:\$PATH" >> ~/.bash_profile $ source ~/.bash_profile # Test installation $ cd getcontacts/example/5xnd $ get_dynamic_contacts.py --topology 5xnd_topology.pdb \ --trajectory 5xnd_trajectory.dcd \ --itypes hb \ --output 5xnd_hbonds.tsv
# Make sure you have git and conda installed and then run # Install get_contact_ticc.py dependencies conda install scipy numpy scikit-learn matplotlib pandas cython pip install ticc==0.1.4 # Set up vmd-python library conda install -c https://conda.anaconda.org/rbetz vmd-python # Set up getcontacts library git clone https://github.com/getcontacts/getcontacts.git echo "export PATH=`pwd`/getcontacts:\$PATH" >> ~/.bashrc source ~/.bashrc
-
N.B. DGLLife requires
rdkit==2018.09.3conda install biopython conda install -c conda-forge rdkit==2018.09.3
-
Install DSSP:
We use DSSP for computing some protein features
$ conda install -c salilab dssp
-
Install PyTorch, DGL and DGL LifeSci:
N.B. Make sure to install appropriate version for your CUDA version
# Install PyTorch: MacOS $ conda install pytorch torchvision -c pytorch # Only CPU Build # Install PyTorch: Linux $ conda install pytorch torchvision cpuonly -c pytorch # For CPU Build $ conda install pytorch torchvision cudatoolkit=9.2 -c pytorch # For CUDA 9.2 Build $ conda install pytorch torchvision cudatoolkit=10.1 -c pytorch # For CUDA 10.1 Build $ conda install pytorch torchvision cudatoolkit=10.2 -c pytorch # For CUDA 10.2 Build # Install DGL. N.B. We require 0.4.3 until compatibility with DGL 0.5.0+ is implemented $ pip install dgl==0.4.3 # Install DGL LifeSci $ conda install -c dglteam dgllife
-
Install PyTorch Geometric:
$ pip install torch-scatter==latest+${CUDA} -f https://pytorch-geometric.com/whl/torch-${TORCH}.html $ pip install torch-sparse==latest+${CUDA} -f https://pytorch-geometric.com/whl/torch-${TORCH}.html $ pip install torch-cluster==latest+${CUDA} -f https://pytorch-geometric.com/whl/torch-${TORCH}.html $ pip install torch-spline-conv==latest+${CUDA} -f https://pytorch-geometric.com/whl/torch-${TORCH}.html $ pip install torch-geometric
Where
${CUDA}and${TORCH}should be replaced by your specific CUDA version (cpu,cu92,cu101,cu102) and PyTorch version (1.4.0,1.5.0,1.6.0), respectivelyN.B. Follow the instructions in the Torch-Geometric Docs to install the versions appropriate to your CUDA version.
-
$ conda install -c schrodinger pymol $ git clone https://github.com/cxhernandez/ipymol $ cd ipymol $ pip install .
N.B. The PyPi package seems to be behind the github repo. We require functionality that is not present in the PyPi package in order to construct meshes.
-
Install graphein:
$ git clone https://www.github.com/a-r-j/graphein $ cd graphein $ pip install -e .
Please consider citing graphein if it proves useful in your work.
@article{Jamasb2020,
doi = {10.1101/2020.07.15.204701},
url = {https://doi.org/10.1101/2020.07.15.204701},
year = {2020},
month = jul,
publisher = {Cold Spring Harbor Laboratory},
author = {Arian Rokkum Jamasb and Pietro Lio and Tom Blundell},
title = {Graphein - a Python Library for Geometric Deep Learning and Network Analysis on Protein Structures}
}


