
IPython Extension for semantic web technology support (Turtle, SPARQL, ShEx, etc.)
This extension is meant to be used together with Jupyter Notebooks for educational purposes. Its focus is neither performance nor scalability but instead ease-of-use.
Jupyter-RDFify was developed by the Chair of Information Systems at RWTH Aachen University to support interactive teaching of Semantic Web Technologies. You can find an examplary set of tutorial-like Jupyter Notebooks using Jupyter-RDFify at https://github.com/SemWebNotebooks/Notebooks/tree/main/Notebooks and further information on how to teach Semantic Web Technologies with Jupyter Notebooks, Jupyter-RDFify, the Moodle elearning system and automatic grading at https://github.com/SemWebNotebooks/Notebooks.
DeprecationWarning: The rdflib-jsonld package has been integrated into rdflib as of rdflib==6.0.0. Please remove rdflib-jsonld from your project's dependencies.
You may get this deprecation warning when loading the extension. This is the fault of SPARQLWrapper which jupyter-rdfify depends on. It will stop appearing as soon as SPARQLWrapper removes json-ld from its dependencies. You can safely ignore this warning.
python -m pip install jupyter-rdfify
python -m pip install git+https://github.com/SemWebNotebooks/Jupyter-RDFify
You will need to have Graphviz installed and added to your path.
If you're using Anaconda, you can install the Graphviz binaries using conda:
conda install -c conda-forge graphviz
You first need to use the predefined %load_ext or %reload_ext magic to load Jupyter-RDFify. You need to do this each time your kernel is restarted.
%load_ext jupyter-rdfify
If you've installed the extension correctly, this should register the %rdf magic. This magic is special in that it is interpreted like a command line interface. If at any point you're wondering what arguments there are and what they do, do not hesitate to use the --help or -h flag.
To list all submodules:
%rdf --help
To list all arguments of the Turtle submodule:
%rdf turtle --help
Jupyter-RDFify is split into several submodules. Select the submodule you want using %rdf <submodule>
These submodules allow you to parse and then visualize or convert graphs. The following graph serialization modules are provided by default: turtle, n3, json-ld, xml
To parse and visualize a graph, just use the %%rdf cell magic with the right submodule. For example to visualize a graph in Turtle notation:
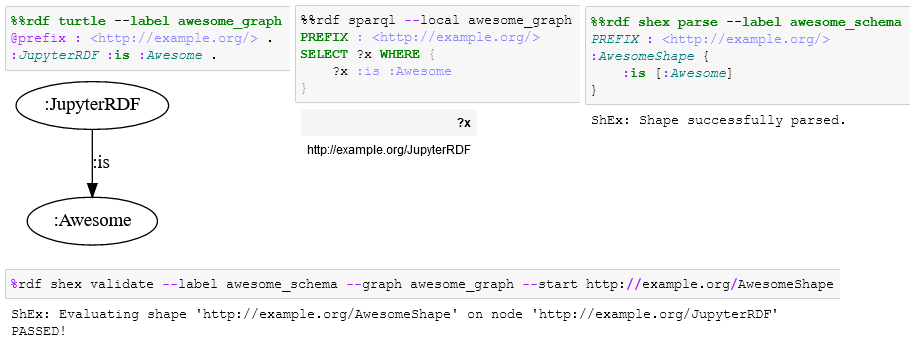
%%rdf turtle
@prefix : <http://example.org/> .
:JupyterRDF :is :Awesome .If this throws an error you probably do not have Graphviz installed. If you do not want to use the visualization, just use --display none or --display table.
To parse and convert a graph into a different format, use a combination of --display raw and --serialize <format>. Possible formats are: turtle, n3, json-ld, xml
To convert a graph in Turtle notation to JSON-LD notation:
%%rdf turtle --display raw --serialize json-ld
@prefix : <http://example.org/> .
:JupyterRDF :is :Awesome .After parsing a graph, you may want to give it a label. You can later use this label to reference your graph in other submodules. With this you can for instance query, validate, draw or entail your graph later on. To give your graph a label just use the --label <label> or -l <label> argument.
%%rdf turtle --label awesome_graph
@prefix : <http://example.org/> .
:JupyterRDF :is :Awesome .The special label last will always hold the last object, even if no --label argument was supplied.
You can use the SPARQL submodule to query existing endpoints or to query local graphs.
Use the --endpoint argument to query an endpoint. An example using the Wikidata endpoint:
%%rdf sparql --endpoint https://query.wikidata.org/sparql
SELECT ?item ?itemLabel
WHERE
{
?item wdt:P31 wd:Q146.
SERVICE wikibase:label { bd:serviceParam wikibase:language "[AUTO_LANGUAGE],en". }
} LIMIT 10You can query labelled graphs using the --local <label> argument. Note that this overrides the endpoint argument.
Example querying the above labelled graph:
%%rdf sparql --local awesome_graph
PREFIX : <http://example.org/>
SELECT ?x WHERE {
?x :is :Awesome
}With this submodule, you can validate graphs using the ShEx language. You first need to parse a schema:
%%rdf shex parse --label awesome_schema
PREFIX : <http://example.org/>
:AwesomeShape {
:is [:Awesome]
}
We already gave the schema a label to reference it later. Note that graph labels and schema labels use different namespaces and do not overwrite each other.
To validate a graph, we need to provide the schema label (--label), the graph label (--graph) of the graph we want to validate against the schema and at least a starting shape (--start). This will check every node of the graph against the starting shape. If we want to focus on a specific node in the graph, we can use the --focus argument. To validate the awesome_graph against the awesome_schema, starting from the :AwesomeShape and focussing on the :JupyterRDF node:
%rdf shex validate --label awesome_schema --graph awesome_graph --start http://example.org/AwesomeShape --focus http://example.org/JupyterRDF
As many of the languages/formats use prefix declarations and these usually just distract from the actual task, most submodules lets you outsource them. Using either the --prefix flag or the prefix action (only for ShEx submodule), you can define a string which gets prepend to every cell magic of that submodule. A simple example for Turtle which defines some very frequent prefixes:
%rdf turtle --prefix
@prefix rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#> .
@prefix rdfs: <http://www.w3.org/2000/01/rdf-schema#> .
@prefix xsd: <http://www.w3.org/2001/XMLSchema#> .The graph manager submodule lets you list, draw, entail and delete labelled graphs. You just need to specify the action and usually a graph label. To draw or awesome_graph:
%rdf graph draw --label awesome_graph
Using OWL-RL, you can generate the finite closure of a graph under either RDFS semantics, OWL-RL semantics or both. This uses a brute-force approach, so it may easily take a long time or fail for large graphs. You can either entail a parsed graph directly using the --entail <regime> argument or entail a labelled graph later using the graph manager:
%rdf graph entail-<regime> --label awesome_graph
For now you can only entail graphs in-place. Possible values for <regime>: rdfs, owl, rdfs+owl
Note that these dependencies will be installed automatically if you use Pip.
RDFLib: The heart of this extension
RDFLib-jsonld: Extension of RDFLib for JSON-LD
SPARQLWrapper: Extension of RDFLib for SPARQL
OWL-RL: Library for RDFS and OWL-RL entailment
PyShEx: Implementation of ShEx
Graphviz python wrapper
IPython