TODO make code better make outputs nicer make input from vcf file
Some of the worst code I've ever written, but the results table is what I intended. The graphs arent great and the code really is terrible.
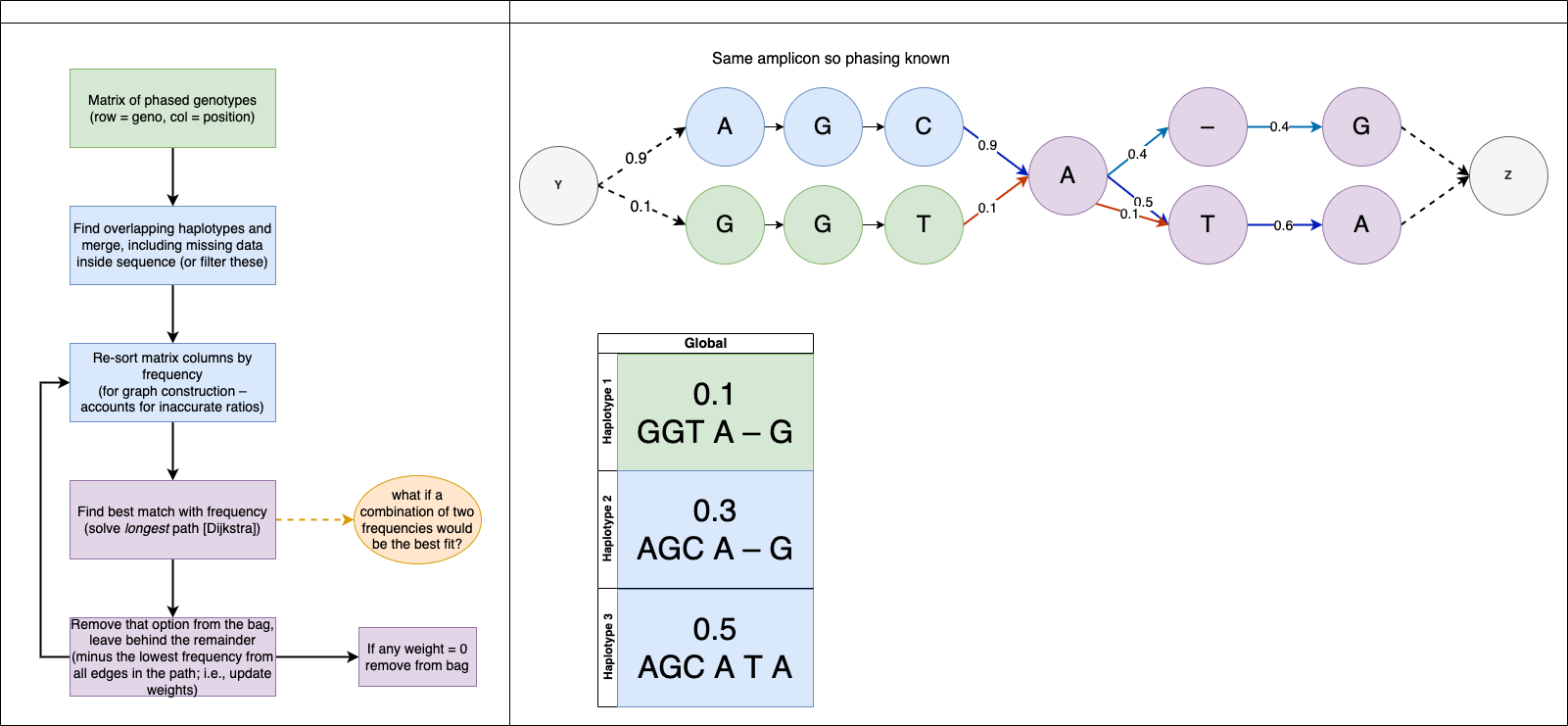
This takes in two files, a bam alignment of short read amplicons and a table of the identified mutations and attempts to determine the most probably haplotypes.
- DT
- visNetwork
- igraph
- stringr
- tidyverse
- reticulate
conda env create -n hasan --file hasan_conda.yml
''' bash run_haplo.sh "path to bam file" "path to snps file" '''
| CHROM | POS | REF | ALT | TYPE |
|---|---|---|---|---|
| rv0678_500 | 489 | C | A | SNP |
| rv0678_500 | 489 | C | C | SNP |
| rv0678_500 | 874 | T | C | SNP |
| rv0678_500 | 874 | T | T | SNP |