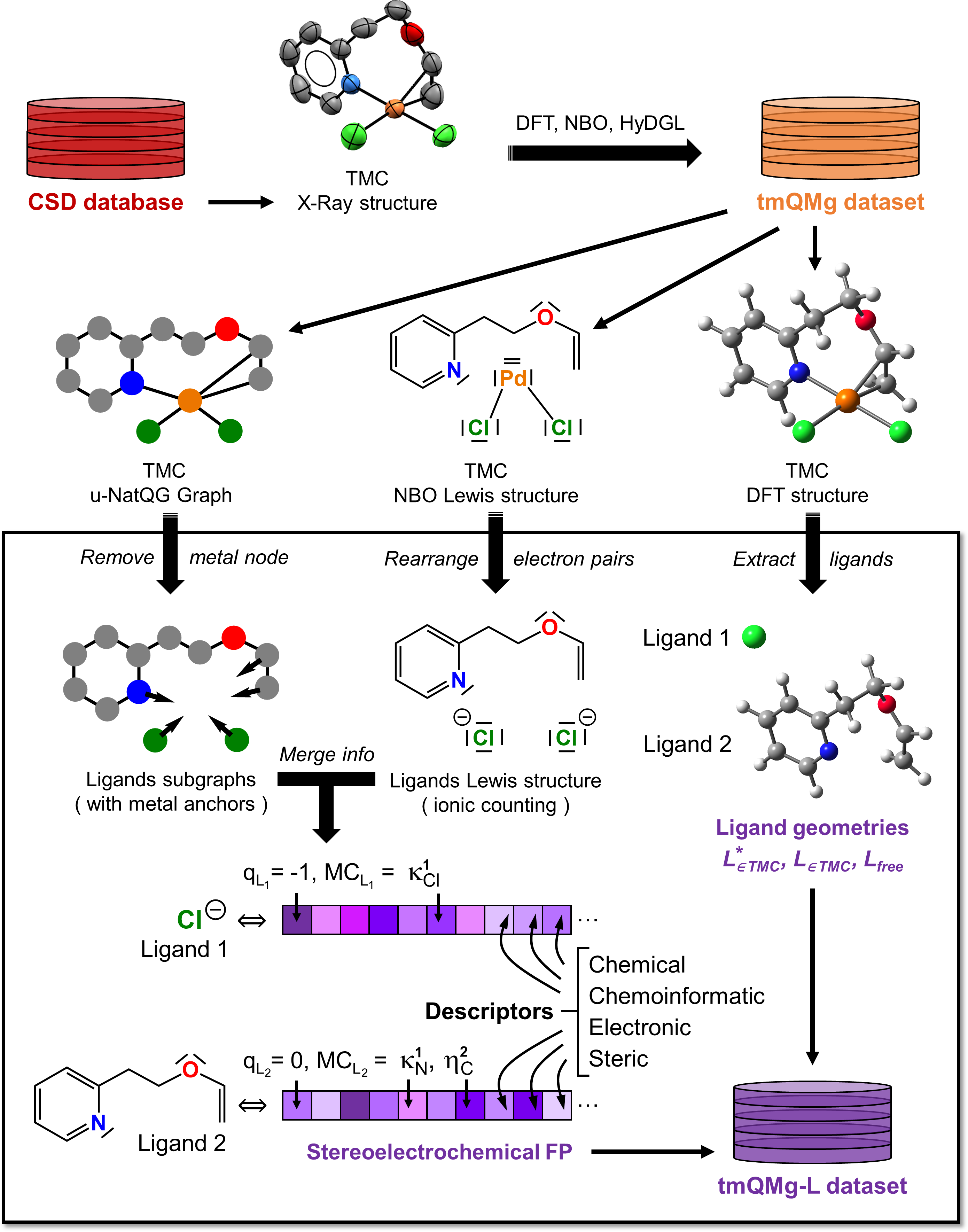
This repository contains the data files of the ligands dataset tmQMg-L containing 29k ligands extracted from the Cambridge Structural Database. The ligands come with their atomic positions, metal-coordinating atom indices and corresponding formal charges. Electronic, steric and cheminformatics descriptors have been calculated for each ligand and are included as well. Details on how the data was compiled can be found in the corresponding publication Directional Multiobjective Optimization of Metal Complexes at the Billion-Scale with the tmQMg-L Dataset and PL-MOGA Algorithm.
- The main ligand file containing information about IDs, SMILES, stoichiometry, occurrence and metal-coordinating atom indices.
- The column
parent_metal_occurrencescontains for all ligands a serialized Python dictionary with the different metal elements as keys and lists of occurrence names as values. For example, an entry might read{'Ni': ['XXYYZZ-subgraph-0', 'XXYYZZ-subgraph-2'], 'Pt': ['ZZYYXX-subgraph-0']}, which would mean that this particular ligand has in total three occurrences where two of them come from the TMC with CSD code XXYYZZ with Nickel as the metal center and one of them comes from the TMC with CSD code ZZYYXX with Platinum as the metal center. - The column
smiles_metal_bond_node_idx_groupscontains for all ligands the indices of metal coordinating atoms in the corresponding SMILES string. Note that they are stored as a list of lists to denote denticity and hapticity. Metal coordinating atom indices in one sublist are haptic meaning that they are continuous in the ligand whereas indices in different sublists are dentic with respect to each other. - The column
metal_bond_node_idx_groupscontains for all ligands a serialized Python dictionary with the different occurrence names as keys and the corresponding metal coordinating atom indices as values. This distinction is necessary because the ordering of atoms in the xyz files of different occurrences of the same ligand differ. For example, an entry might read{'XXYYZZ-subgraph-0': [[0], [2,3]], 'XXYYZZ-subgraph-2': [[1],[3,5]], 'ZZYYXX-subgraph-0': [[5],[1,4]]}which would mean that this particular ligand has three occurrences, each with different metal coordinating atom indices due to different ordering of atoms in the xyzs. Note again that the coordinating atom indices are stored as a list of lists to denote denticity and hapticity. Metal coordinating atom indices in one sublist are haptic meaning that they are continuous in the ligand whereas indices in different sublists are dentic with respect to each other.
- The ligand fingerprints for all ligands containing information such as the charge, number of atoms and their type.
- The column
chargecontains the NBO derived formal charge as described in the publication. - The columns
n_metal_bound,n_dentic_bound, andn_haptic_boundrefer to the number of atoms bound to the metal center, the number of which are dentic, and the number of which are haptic, respectively. - The column
n_atomscontains the total number of atoms in the ligand. For each element, the number of occurrences in the ligand is listed in columnXwhere X denotes the element symbol. - The columns
dentic_Xandhaptic_Xwhere X denotes the element symbol contain the number of dentic/haptic bound atoms of that element. - The column
is_alternative_chargeis a boolean flag that denotes if a ligand is occuring with an alternative charge. For some of the ligands, the charge determination algorithm gave multiple different charges for different occurrences of the same ligand (same topology and connection atoms). Usually one charge was in the majority and all others were discarded as outliers/errors. However, in cases where another charge was present in a significant amount (>25%) it was also recorded with the flagis_alternative_chargeset toTrue.
- The calculated RDKit, steric and electronic descriptors for all ligands as described in the publication.
- Some properties were specifically calculated based on either the geometry of the most stable occurrence or for the gas phase optimized structure. The column names reflect this with the prefix
L*to refer to the most stable occurrence and the prefixL_freeto refer to the relaxed structure. Properties without a prefix were simply calculated based on the ligands SMILES string.
- List of all ligands and their most stable occurrence.
- List of Weisfeiler-Lehman graph hashes for all ligands.
- The column
base_hashrefers to the hashes only considering connectivity. - The columns
atom_attribution_hash,bond_attribution_hash, andatom_bond_attribution_hashrefer to the hashes also considering atom element, bond order, and both atom element as well as bond order, respectively.
- A list of ligands included in the OctLig (Kulik and co-workers, DOI: 10.1021/acs.jctc.2c00468) and tmQMg-L datasets and their determined charges. The columns
OctLig_nameandtmQMg-L_nameprovide the identifiers from both the OctLig and tmQMg-L datasets, respectively, and the columnsOctLig_chargeandtmQMg-L_chargeprovide their corresponding predicted charges. The columncharge_agreementcontains TRUE if the two charges agree and FALSE otherwise. The last column provides the SMILES string for each ligand. Graph matching was done by generating cutoff radius graphs for all ligands of both datasets and performing node attributed graph isomorphy tests to determine equivalent ligands in the two datasets. If a ligand is only found in the OctLig dataset but not in tmQMg-L, only the columnsOctLig_nameandsmileswill contain entries.
- Directory containing the RDKit, steric and electronic descriptors for all ligands in separate files, the scripts to create them, and a script to merge them into one.
- Directory containing the geometries of all ligands (xyz/ligands_xyzs.xyz), only the stable ligands (xyz/ligands_stable_xyzs.xyz) and the optimized stable ligands (xyz/ligands_stable_xyzs_opt.xyz).
- With Python the xyzs can easily be loaded as a dictionary with the occurrence names as keys and the xyzs as values using the following code snippet:
xyzs = {}
with open('./xyz/ligands_xyzs.xyz)', 'r') as fh:
for xyz in fh.read().split('\n\n'):
xyzs[xyz.split('\n')[1]] = xyz
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.