Accepted in NeurIPS Workshop 2023 (AI4D3 | New Frontiers of AI for Drug Discovery and Development) [arxiv]
Official Github for PharmacoNet: Accelerating Large-Scale Virtual Screening by Deep Pharmacophore Modeling by Seonghwan Seo* and Woo Youn Kim.
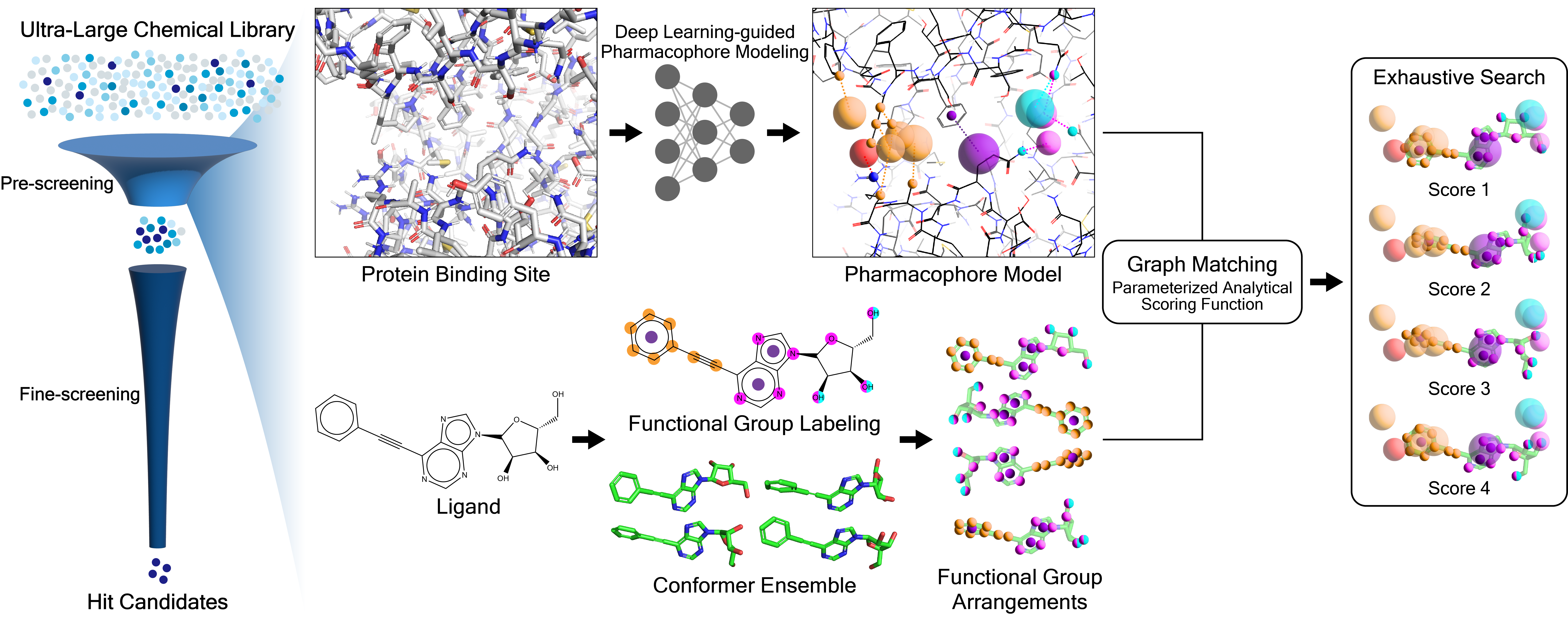
- Fully automated protein-based pharmacophore modeling based on image instance segmentation modeling
- Coarse-grained graph matching at the pharmacophore level
PharmacoNet is an extremely rapid yet reasonably accurate structure-based scoring function with high generation ability.
If you have any problems or need help with the code, please add an issue or contact shwan0106@kaist.ac.kr.
# Download Pre-trained Model
sh ./download-weights.sh
# Pharmacophore Modeling
python modeling.py -r <RECEPTOR_PATH> --autobox_ligand <LIGAND_PATH> -p <OUT_PHARMACOPHORE_MODEL_PATH>
python modeling.py -r <RECEPTOR_PATH> --center <X> <Y> <Z> -p <OUT_PHARMACOPHORE_MODEL_PATH> --cuda # CUDA Acceleration
# Scoring with a single ligand SMILES
python scoring.py -p <PHARMACOPHORE_MODEL_PATH> -s <SMILES> --num_conformers <NUM_CONFORMERS>
# Scoring with a ligand SMILES file. (see ./examples/example.smi)
python scoring_file.py -p <PHARMACOPHORE_MODEL_PATH> -s <SMILES_FILE> --num_conformers <NUM_CONFORMERS> --num_cpus <NUM_CPU># Pharmacophore Modeling for KRAS(G12C) - PDBID: 6OIM
python modeling.py -r ./examples/6OIM_protein.pdb --autobox_ligand ./examples/6OIM_ligand.sdf -p ./examples/6OIM_model.json
# Scoring
python scoring.py -p ./examples/6OIM_model.json -s 'c1ccccc1' --num_conformers 8 # 8 is arbitrary value.
python scoring_file.py -p ./examples/6OIM_model.json -s ./examples/example.smi --num_conformers 10 --num_cpu 8# Required python>=3.9, Best Performance at higher version. (3.9, 3.10, 3.11, 3.12 - best)
conda create --name pmnet python=3.11
conda activate pmnet
conda install openbabel
pip install torch torchvision # torch >= 1.13, CUDA acceleration is available. 1min for 1 cpu, 10s for 1 gpu
pip install rdkit biopython omegaconf timm numba # Numba is optional, but recommended.
pip install molvoxel # https://github.com/SeonghwanSeo/molvoxel.gitWe provide two simple example scripts for scoring. However, it can be easily included in your custom script via the python code below.
* Multiprocessing is allowed. See ./scoring_file.py
from src import PharmacophoreModel
from openbabel import pybel
model = PharmacophoreModel.load(<PHARMCOPHORE_MODEL_PATH>)
pbmol = pybel.readstring('smi', <LIGAND_SMILES>)
# NOTE: Scoring with RDKit ETKDG Conformers
def scoring_etkdg_conformers(model, pbmol, smiles, num_conformers):
from rdkit import Chem
from rdkit.Chem import AllChem
rdmol = Chem.MolFromSmiles(smiles)
rdmol = Chem.AddHs(rdmol)
AllChem.EmbedMultipleConfs(rdmol, num_conformers, AllChem.srETKDGv3())
rdmol = Chem.RemoveHs(rdmol)
return model.scoring(pbmol, rdmol)
# NOTE: Scoring with custom ligand conformers.
def scoring_own_conformers(model, pbmol, atom_positions):
# atom_positions: [NUM_CONFORMERS, NUM_ATOMS, 3]
return model.scoring(pbmol, atom_positions=atom_positions)Paper on arxiv
@article{seo2023pharmaconet,
title = {PharmacoNet: Accelerating Large-Scale Virtual Screening by Deep Pharmacophore Modeling},
author = {Seo, Seonghwan and Kim, Woo Youn},
journal = {arXiv preprint arXiv:2310.00681},
year = {2023},
url = {https://arxiv.org/abs/2310.00681},
}