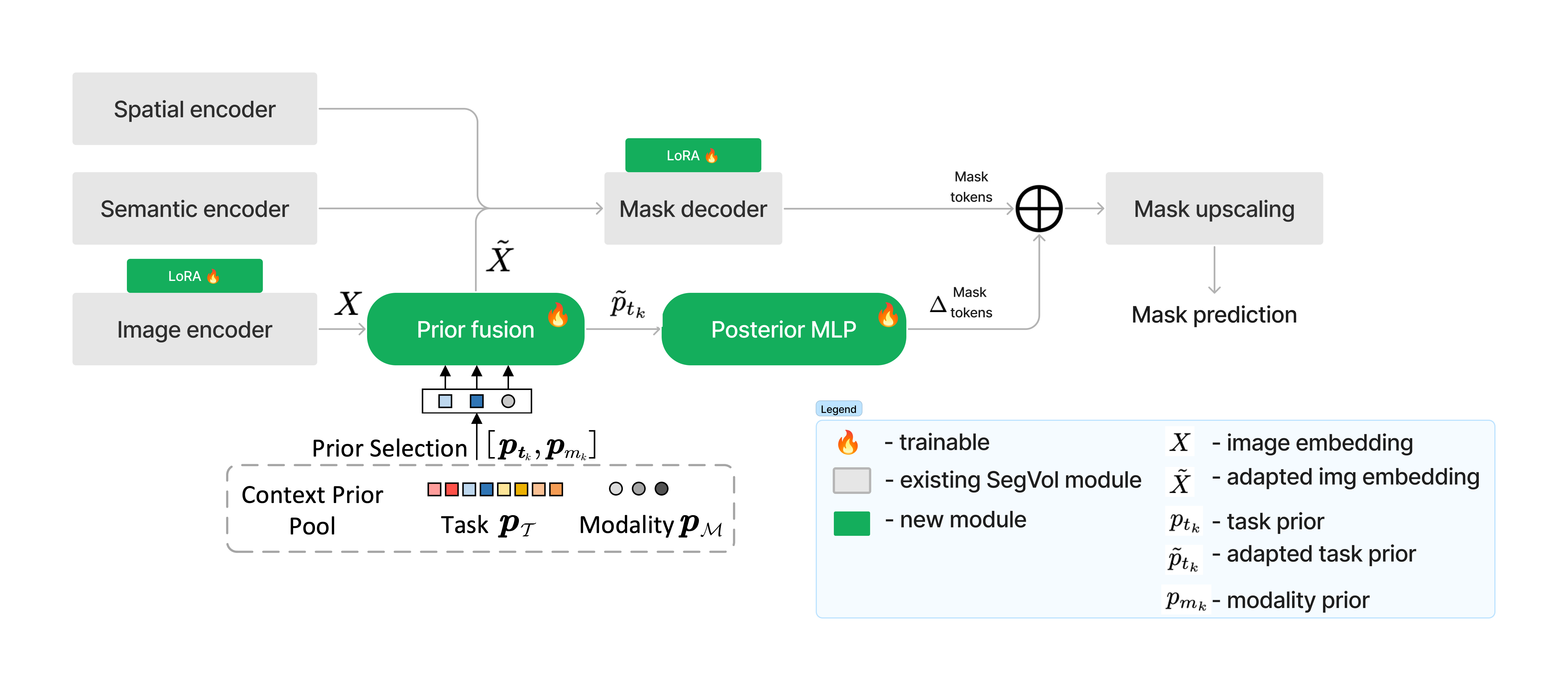
Keywords: 3D medical SAM, volumetric image segmentation, LoRA, Context-Prior Learning
This repository contains a reproduction and extension of "SegVol: Universal and Interactive Volumetric Medical Image Segmentation" by Du et al. (2023) using LoRA adapters and context priors introduced in "Training Like a Medical Resident: Context-Prior Learning Toward Universal Medical Image Segmentation" by Gao et al. (2023)
To read the full report containing detailed information on our experiments and extension study, please, refer to our blogpost.
To get started, clone the repository and install the dependencies using Poetry.
-
Clone the environment
git clone https://github.com/SergheiMihailov/adapt_med_seg.git
-
Activate the existing Poetry environment:
poetry shell
-
Install the project dependencies (if not already installed):
poetry install
This will activate the primary environment with all necessary dependencies for the main functionalities of this project.
This project uses the M3D-Seg dataset, which contains 25 datasets involving various medical imaging modalities such as CT of different anatomical structures. The 25 processed datasets are being uploaded to ModelScope and HuggingFace. Additionally, we augment this dataset with the following datasets:
| Dataset | Modality | Link |
|---|---|---|
| AMOS_2022 | CT, MRI | https://amos22.grand-challenge.org/ |
| BRATS2021 | MRI | https://www.med.upenn.edu/cbica/brats2021/ |
| CHAOS | CT, MRI | https://chaos.grand-challenge.org/ |
| MSD | CT, MRI | http://medicaldecathlon.com/ |
| SAML_mr_42 | MRI | https://kaggle.com/datasets/nguyenhoainam27/saml-mr-42 |
| T2-weighted-MRI | MRI | https://kaggle.com/datasets/nguyenhoainam27/t2-weighted-mri |
| promise12_mr | MRI | https://promise12.grand-challenge.org/ |
Due to resources constraints, the subsamples of the above datasets are used for training and evaluation. These subdatasets were sampled by taking one random image from the first dataset, then from the second etc. until (200,400,800) reached, or there was no more unique sample in the specific dataset.
| Dataset Size (Num. Samples) | Download Link |
|---|---|
| 200 | Download |
| 400 | Download |
| 800 | Download |
Each of these datasets contain the following structure
datasets/
├── M3D_Seg/
├── AMOS_2022/
├── BRATS2021/
├── CHAOS/
├── MSD/
├── SAML_mr_42/
├── T2-weighted-MRI/
└── promise12_mr/We have trained two configurations on top of the SegVol model. These checkpoints, along with the base model from the SegVol paper, can be found below:
| Model name | Dice score(%) | Download Checkpoint |
|---|---|---|
| segvol_baseline | 0.5744 (0.70691) | Download |
| segvol_lora | 0.6470 | Download |
| segvol_context_prior | 0.6651 | Download |
The training pipeline is defined in adapt_med_seg/train.py. To train the model, run:
python -m adapt_med_seg.train \
--model_name ${MODEL_NAME} \ # i.e "segvol_baseline"
--dataset_path ${DATASET} \
--modalities CT MRI \
--epochs 10 \
--lora_r 16 \ # Optional: sets the rank of the LoRA adapter.
--lora_alpha 16 # Optional: sets the alpha value for the LoRA adapter.The evaluation pipeline is defined in adapt_med_seg/pipelines/evaluate.py. To evaluate the model, run:
python -m adapt_med_seg.eval \
--model_name ${MODEL_NAME} \ # i.e "segvol_baseline"
--dataset_path ${DATASET} \
--modalities CT MRI \
--ckpt_path ${CHECKPOINT_PATH} \ i.e "segvol_lora.ckpt"
--lora_r 16 \ # Optional but need to match the training lora_r of the checkpoint: sets the rank of the LoRA adapter.
--lora_alpha 16 \ # Optional but need to match the training lora_rof the checkpoint: sets the alpha value for the LoRA adapter.This section provides an overview of the available Jupyter notebooks designed to help you with various tasks such as preprocessing data, performing inference, and visualizing results.
Several Jupyter notebooks are provided for various tasks such as preprocessing, inference, and visualization:
notebooks/inference_colab.ipynb: Inference on Colab.notebooks/inference.ipynb: General inference notebook.notebooks/preprocess.ipynb: Data preprocessing steps.notebooks/process_results.ipynb: Processing and analyzing results.notebooks/SegVol_initial_tryouts.ipynb: Initial segmentation volume tryouts.notebooks/vis.ipynb: Visualization of results.notebooks/zsombor_balance_amos.py: Script for balancing AMOS dataset.- Training notebook: Colab notebook for running model training.
To run these notebooks, activate the Poetry environment and start Jupyter Notebook:
poetry shell
jupyter notebook
|
|
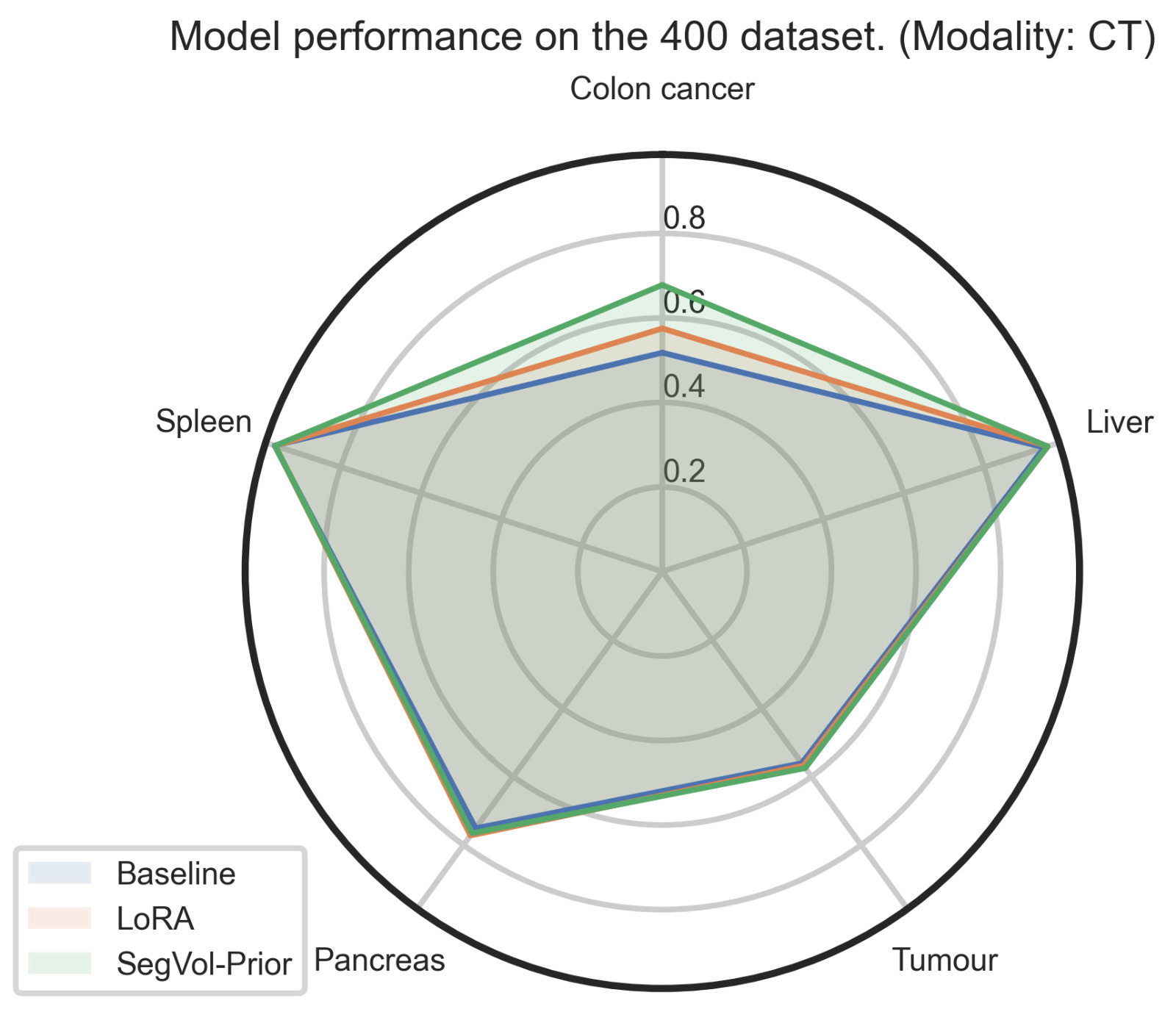
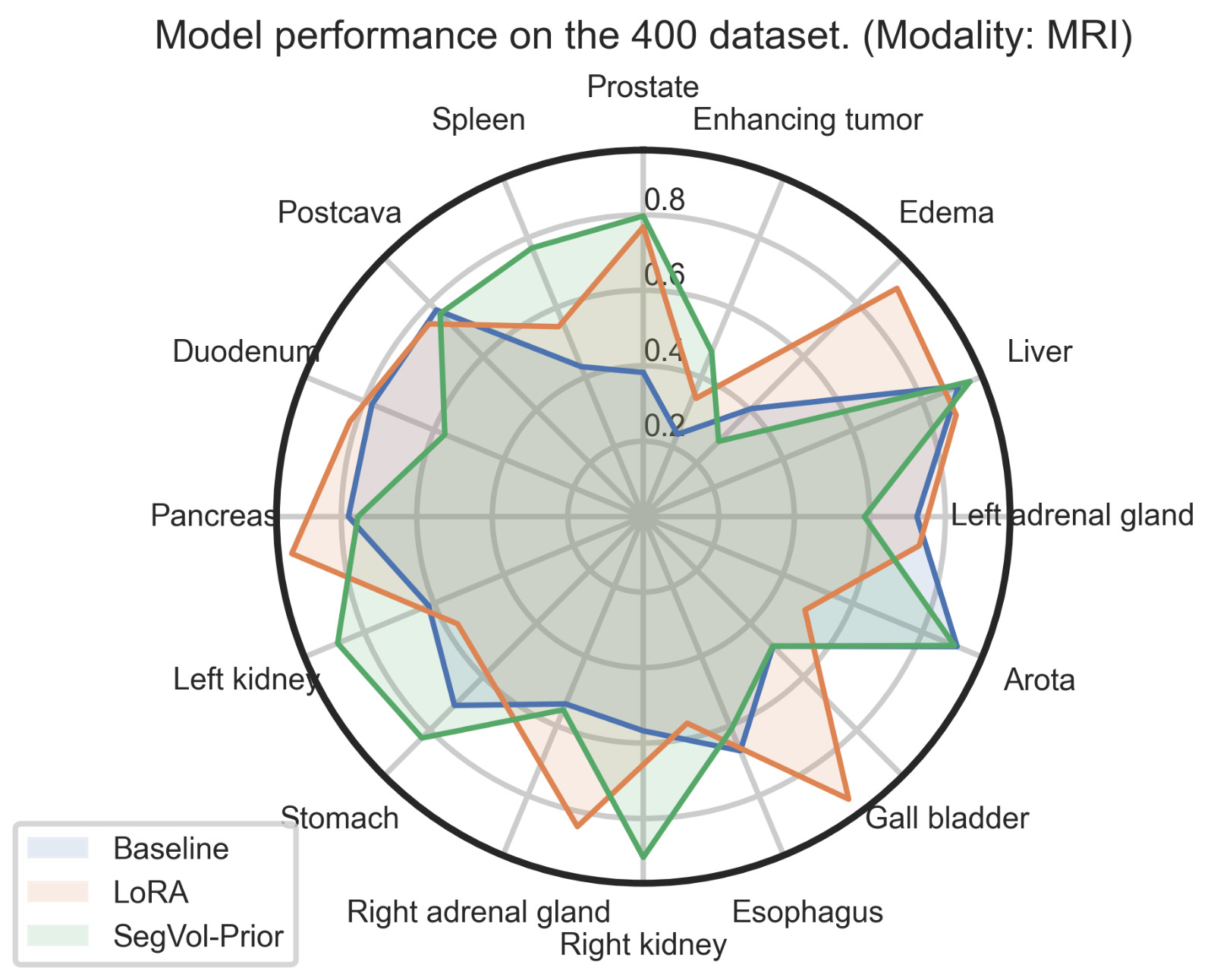
| Figure 4. Combined view of our results over different modalities and organs. | |
If you find this repository helpful, please consider citing:
@misc{SegEVOLution2024,
title = {SegEVOLution: Enhanced Medical Image Segmentation with Multimodality Learning},
author = {Zsombor, Fülöp and Serghei, Mihailov and Matey, Krastev and Miklos, Hamar and Danilo, Toapanta, Stefanos, Achlatis},
year = {2024},
howpublished = {\url{https://github.com/SergheiMihailov/adapt_med_seg.git}},
}
Thanks for the following amazing works: SegVol
CLIP.
Footnotes
-
CT only, our measurement ↩