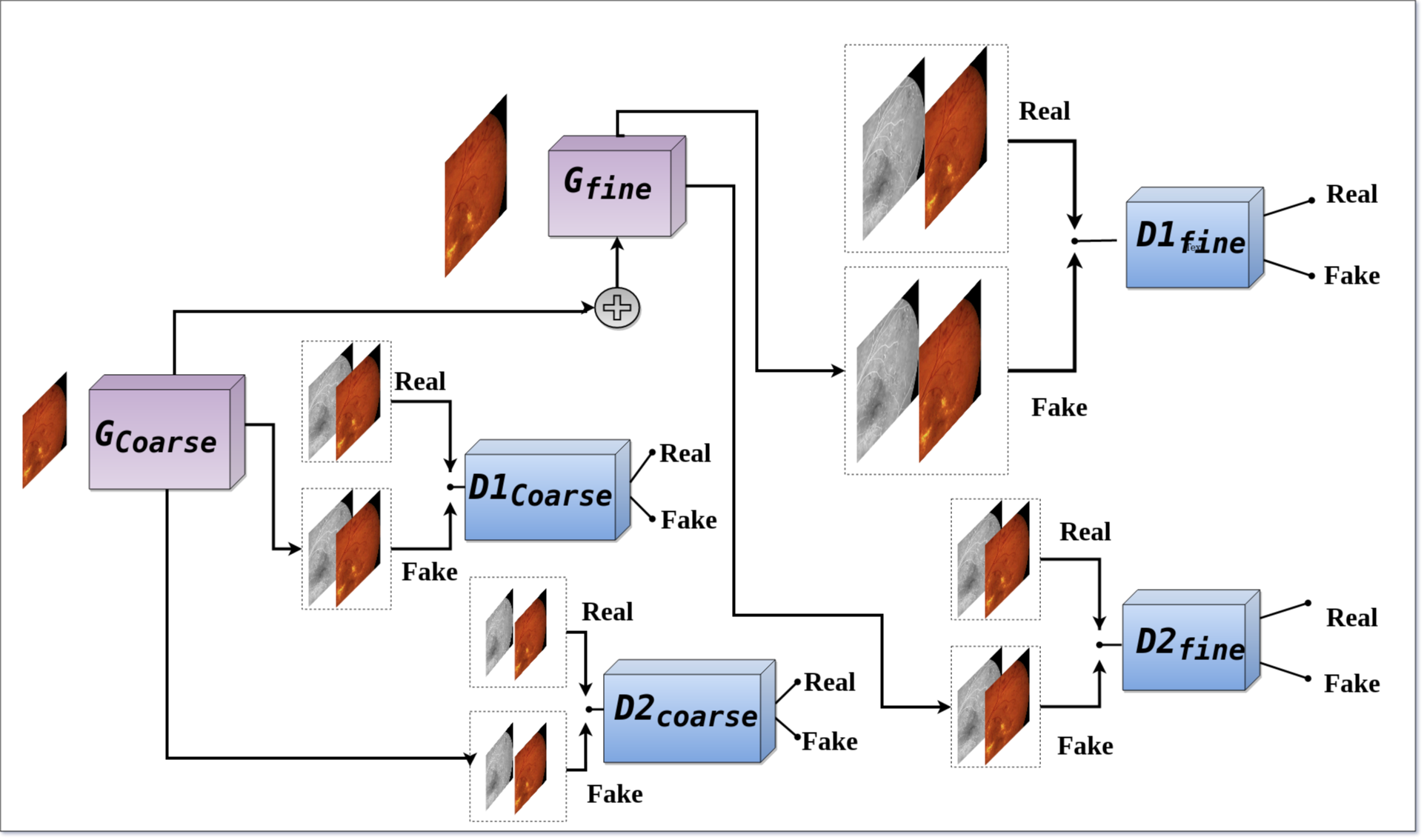
This code is part of the supplementary materials for the ISVC 2020 for our paper Fundus2Angio: A Conditional GAN Architecture for Generating Fluorescein Angiography Images from Retinal Fundus Photography . The paper has since been accpeted to ISVC 2020 and will be preseneted in October 2020.
https://arxiv.org/abs/2005.05267
@inproceedings{kamran2020fundus2angio,
title={Fundus2Angio: A Conditional GAN Architecture for Generating Fluorescein Angiography Images from Retinal Fundus Photography},
author={Kamran, Sharif Amit and Hossain, Khondker Fariha and Tavakkoli, Alireza and Zuckerbrod, Stewart and Baker, Salah A and Sanders, Kenton M},
booktitle={International Symposium on Visual Computing},
pages={125--138},
year={2020},
organization={Springer}
}
- Ubuntu 18.04 / Windows 7 or later
- NVIDIA Graphics card
- Download and Install Nvidia Drivers
- Download and Install via Runfile Nvidia Cuda Toolkit 10.0
- Download and Install Nvidia CuDNN 7.6.5 or later
- Install Pip3 and Python3 enviornment
sudo apt-get install pip3 python3-dev
- Install Tensorflow-Gpu version-2.0.0 and Keras version-2.3.1
sudo pip3 install tensorflow-gpu==2.0.3
sudo pip3 install keras==2.3.1
- Install packages from requirements.txt
sudo pip3 -r requirements.txt
https://sites.google.com/site/hosseinrabbanikhorasgani/datasets-1/fundus-fluorescein-angiogram-photographs--colour-fundus-images-of-diabetic-patients
- Please cite the paper if you use their data
@article{hajeb2012diabetic,
title={Diabetic retinopathy grading by digital curvelet transform},
author={Hajeb Mohammad Alipour, Shirin and Rabbani, Hossein and Akhlaghi, Mohammad Reza},
journal={Computational and mathematical methods in medicine},
volume={2012},
year={2012},
publisher={Hindawi}
}
- Folder structure for data pre-processing given below. Please make sure it matches with your local repository.
├── Dataset
| ├──ABNORMAL
| ├──NORMAL
- Type this in terminal to run the random_crop.py file
python3 random_crop.py --output_dir=data --input_dim=512 --datadir=Dataset
- There are different flags to choose from. Not all of them are mandatory.
'--input_dim', type=int, default=512
'--n_crops', type=int, default=50
'--datadir', type=str, required=True, help='path/to/data_directory',default='Dataset'
'--output_dir', type=str, default='data'
- Convert all the images to npz format
python3 convert_npz.py --outfile_name=fundus2angio --input_dim=512 --datadir=data --n_crops=50
- There are different flags to choose from. Not all of them are mandatory.
'--input_dim', type=int, default=512
'--n_crops', type=int, default=50
'--datadir', type=str, required=True, help='path/to/data_directory',default='data'
'--outfile_name', type=str, default='fundus2angio'
'--n_images', type=int, default=17
- Type this in terminal to run the train.py file
python3 train.py --npz_file=fundus2angio --batch=4 --epochs=100
- There are different flags to choose from. Not all of them are mandatory
'--npz_file', type=str, default='fundus2angio', help='path/to/npz/file'
'--batch_size', type=int, default=4
'--input_dim', type=int, default=512
'--epochs', type=int, default=100
The code is released under the MIT License, you can read the license file included in the repository for details.