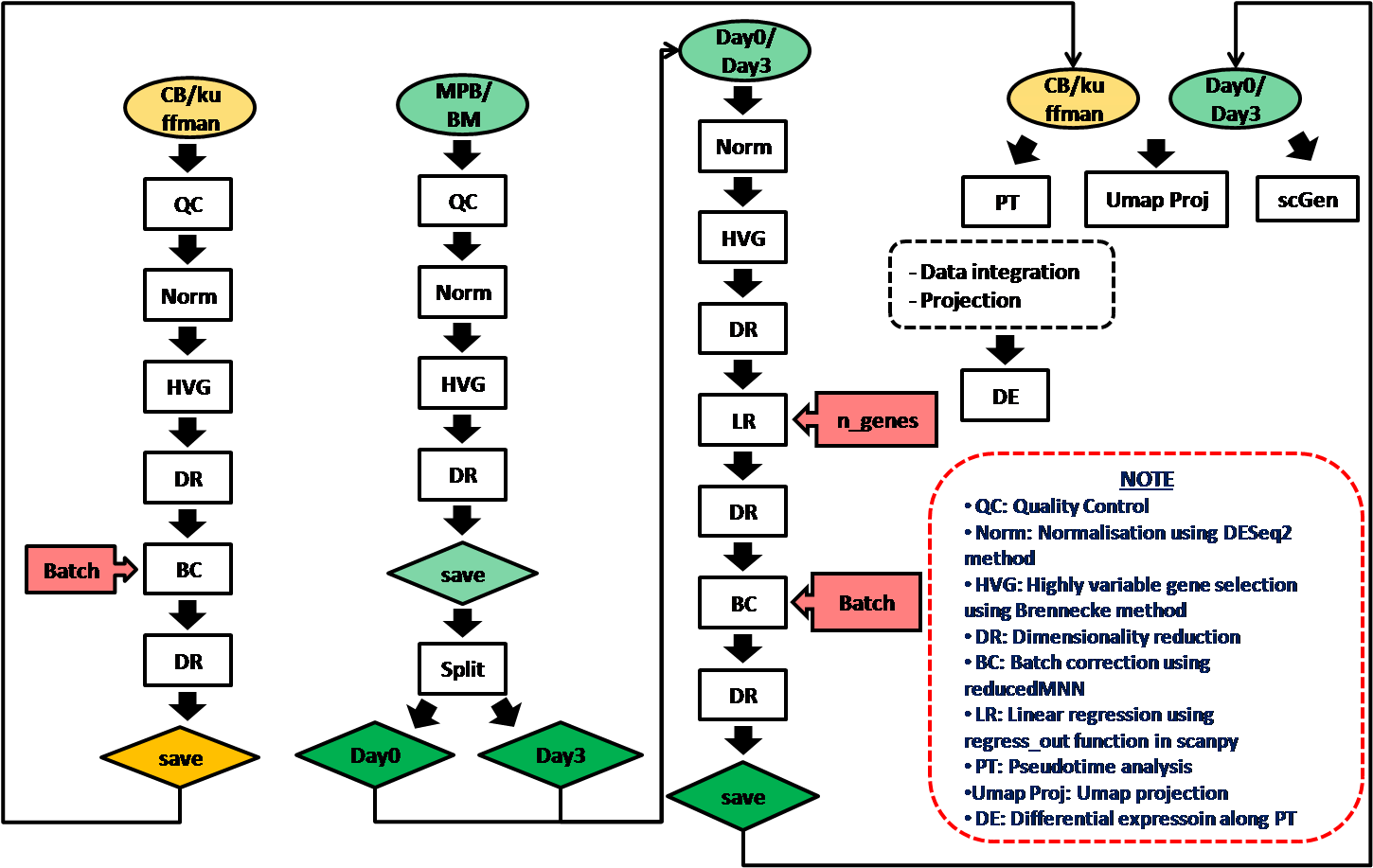
The analysis was done following the strategy below:
Two reference datasets were used for data projection:
- Cord blood dataset (Unpublished)
- Kuffman dataset (Unpublished)
This folder contains all jupyter notebooks that were used for the data analysis. To run the notebooks, the Smart-Seq2 preprocessng package would need to be installed first as
pip install smqpp
The main analysis include:
- CBdata_all: Analysis of Cord blood data to generate visualisation layouts as references for projection.
- Kuffman_all: Analysis of Kuffman data to generate visualisation layouts as references for projection of 0hr data.
- MPB1234_all: Analysis of all MPB cells together, for better batch correction, data was split by days.
- MPB_scGen: Batch correction and prediction of perturbation using scGen for MPB 0hr and 62 hr NT and GFP+ cells.
- MPB1234_Day0: Analysis of MPB 0hr cells
- MPB1234_Day3: Analysis of MPB 62hr cells
- BM789_all: Analylsis of all BM cells together, for better batch correction, data was split by days
- BM_scGen: Batch correction and prediction of perturbation using scGen for BM 0hr and 62 hr NT and GFP+ cells.
- BM789_Day0: Analysis of BM 0hr cells
- BM789_Day3: Analysis of BM 62hr cells
DPT analysis:
- CB test: Check different data integration and projection methods for CB data Batch 1 and Batch2
- Kuffman test: Check different data integration and projection methods for Kuffman data as reference and MPB Day0 and BM Day0
- DI Method1: Data integration and projection for CB data as reference and MPB and BM data using combat for batch correction (BC) following by destiny R package for calculating diffusion map (DM), corresponding projection (Proj) and diffusion pseudotime (DPT)
- DI Method2: Data integration and projection for CB data as reference and MPB and BM data using combat for CB following by scanpy for calculating DM, corresponding Proj and DPT
- DI Method3: Data integration and projection for CB data as reference and MPB and BM data using scanpy for pca calculation whcih were then corrected by reducedMNN in batchelor R package. The corrected PCA was used in scanpy for calculating DM, corresponding Proj and DPT
- Method comparison: Comparing the three methods used for data integration