This repository is the official implementation of Fast abdomen organ and tumor segmentation based-on nnUNet of Team aladdin5 on FLARE23 challenge. Our work heavily based-on the old version nnUNetV2.
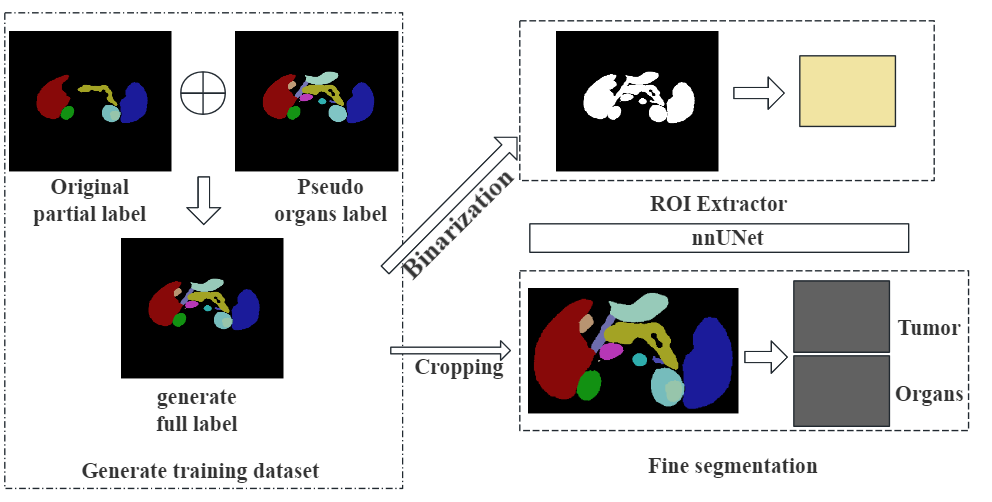
Pipeline of our solutionDetails are presented in the paper. The Training, Inference and Evaluation are scripts for training models, predict samples and calculate the metrics respectively.
- Windows 10
- CPU Intel(R) Core(TM) i7-10700kF CPU@3.80GHz, RAM 16GB x 4, NVIDIA RTX 3090 24G
- CUDA >= 11.1
- python >= 3.8
For training, please follow the install of nnUNet.
The training Data and validation data are provided by the FLARE23. In short, there are 2200 partial labeled and 1800 unlabeled data for training, 100 public cases for validation and 200 hidden cases for the final test. We only use the 2200 partial labeled cases. Besides, this challenge also provide pseudo labels. We make the abdomen ROI dataset by changing the all organs labels to 1 and set the tumors to background.
We follow the preprocessing of nnUnet. For ROI model, we adopt a simple slice method. For fine segmentation, the target spacing for of tumor model and organs model are different. The normalization is same as nnUNet.
Below is the code for preprocessing of ROI extractor, here task_id is the name or id of your specific FLARE23 ROI task
python nnUNet_plan_and_preprocess.py -t Task_id -pl3d ExperimentPlanner3DFabiansResUNet_Varied_Ratio -pl2d None --only_lowres
The preprocessing of fine segmentation is similar, but with different plans.
For tumor fine segmenation
python nnUNet_plan_and_preprocess.py -t Task_id -pl3d ExperimentPlanner3DFabiansResUNet_v21_Tumor -pl2d None
For organs fine segmentation
python nnUNet_plan_and_preprocess.py -t Task_id -pl3d ExperimentPlanner3DFabiansResUNet_v21_Organs -pl2d None
Now you can train your models.
We train the roi model as follows.
python nnunet/run/run_training.py 3d_lowres nnUNetTrainerV2_ResencUNet roi_task_id -f all -p nnUNetPlans_FabiansResUNet_vr
We train the organs model as follow:
python nnunet/run/run_training.py 3d_fullres nnUNetTrainerV2_ResencUNet organs_task_id -f all -p ExperimentPlanner3DFabiansResUNet_v21_Organs
We train the tumor model as follow:
python nnunet/run/run_training.py 3d_fullres nnUNetTrainerV2_ResencUNet_Dynamic_Weight tumor_task_id -f all -p ExperimentPlanner3DFabiansResUNet_v21_Tumor
Besides, we finetuning the tumor model as follow:
python nnunet/run/run_training.py 3d_fullres nnUNetTrainerV2_ResencUNet_finetuning tumor_task_id -f all -p ExperimentPlanner3DFabiansResUNet_v21_Tumor -pretrained_weights path_to_the_previous_tumor_model
You can change the trainer as you need.
After training, we do a lot of work for accelaration, in the end, we can run inference use scripts in folder Inference. You can download our pretrained models Here:
Then put the models into the folder models and run:
python run_inference.py -i path_of_the_input_volumes -o path_for_saving_the_prediction
Maybe you should change the path in run_inferenc(line 141-143).
By the way, we convert the pretrained model into pt format for saving time. So this Inference script can not work for the model trained by nnUNet directly. If you want predict cases with your own models, you should predict in the nnUNet way.
We build our docker under Ubuntu. Here is the example of our tree structure.
/*******/flare23/docker/Dockerfile
/*******/flare23/docker/predict.sh
/*******/flare23/docker/models
/*******/flare23/docker/Inference
/*******/flare23/docker/flare23_all #folder of the all validation images.
/*******/flare23/docker/flare23_test #folder of test images.
Then you can build docker with command below, the pwd is /*******/flare23/docker.
docker build -t aladdin5
To save the docker image
docker save aladdin5 | gzip -c > aladdin5.tar.gz
To load the docker
docker load -i aladdin5.tar.gz
Also, you can download our docker image, with extraction code u12q. The other details are displayed on the FLARE23 Official website..
To compute the evaluation metrics, run FLARE23_DSC_NSD_Eval.py in Evaluation
python FLARE23_DSC_NSD_Eval.py -g ground_truth_path -s save_metric_path -p prediction_path
Then you will get a csv file contains DSC scores and NSD scores of each organ and tumor for every samples.
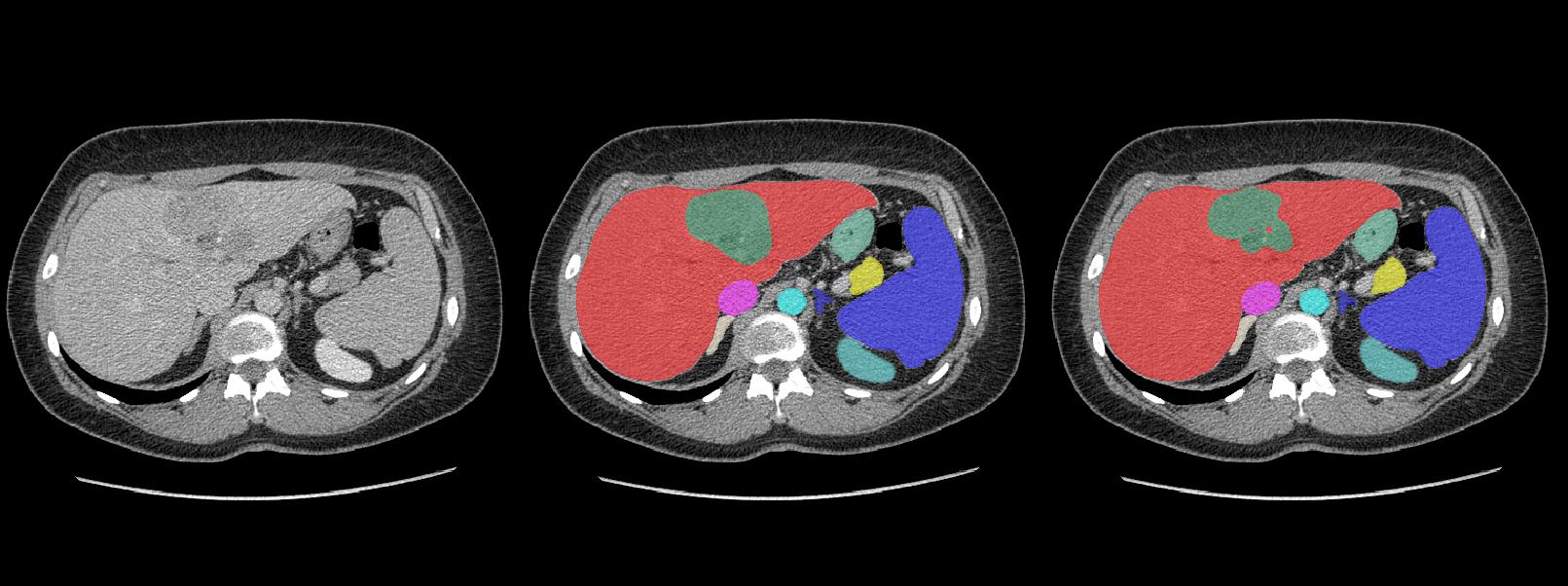
###Well segmented cases
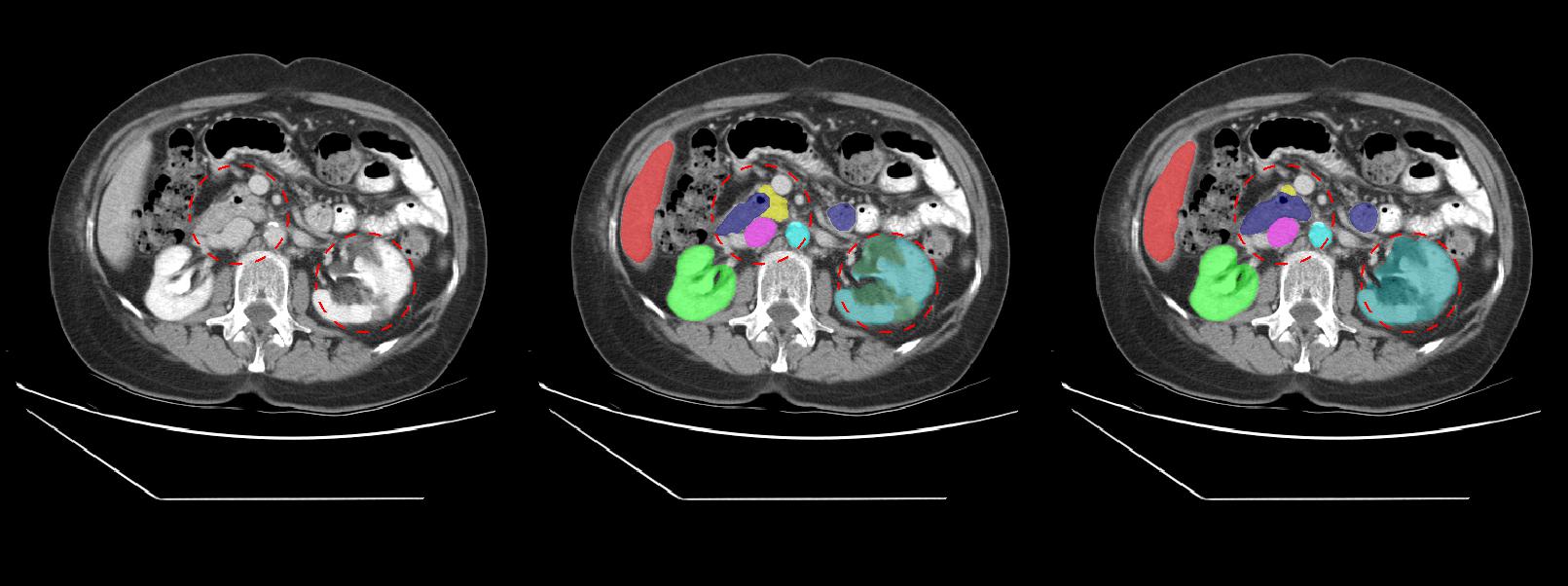
 ###Challenging cases
###Challenging cases
 ###Validation and final test performance
###Validation and final test performance
We thank the contributors of public FLARE23 datasets.