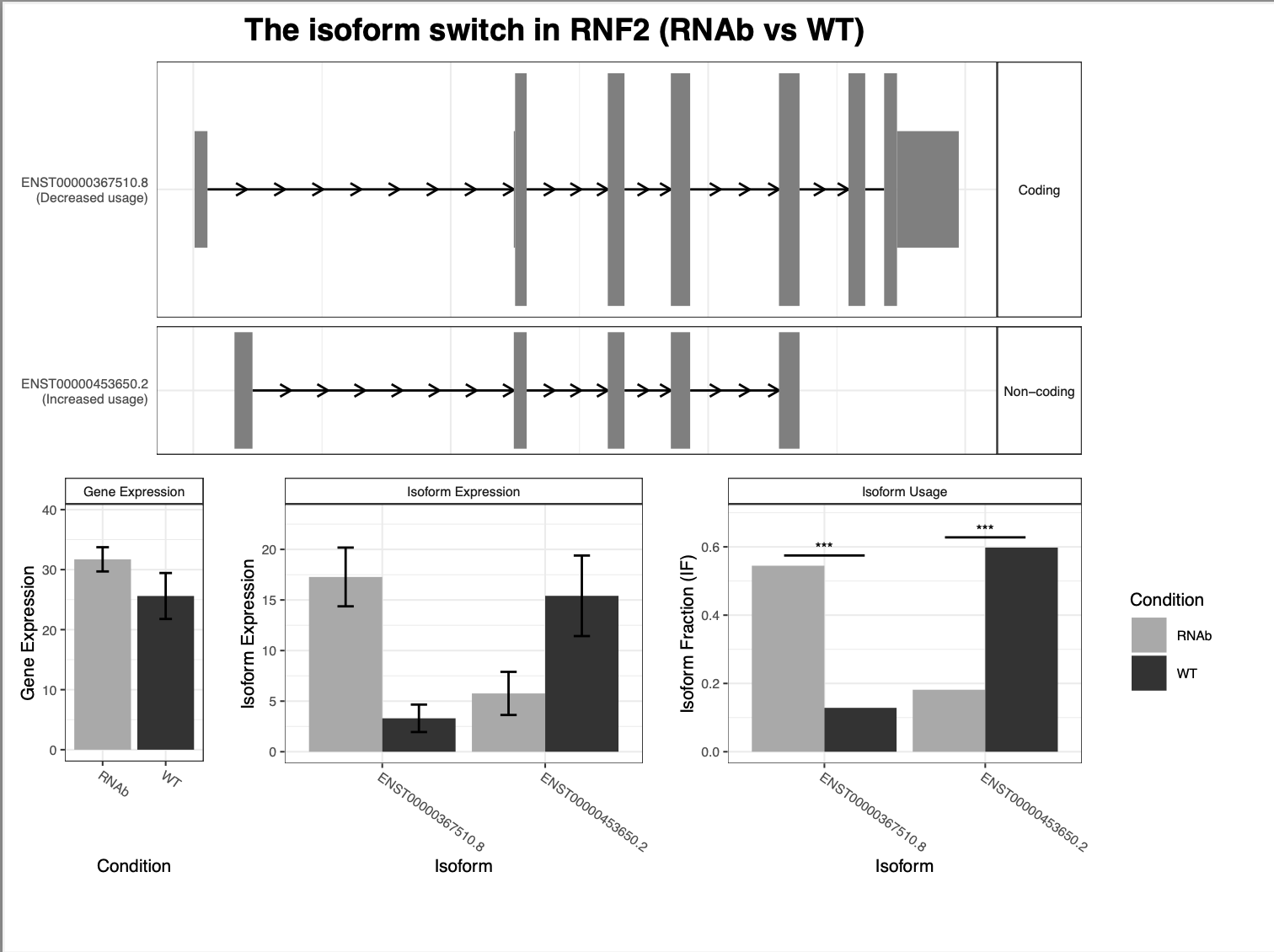
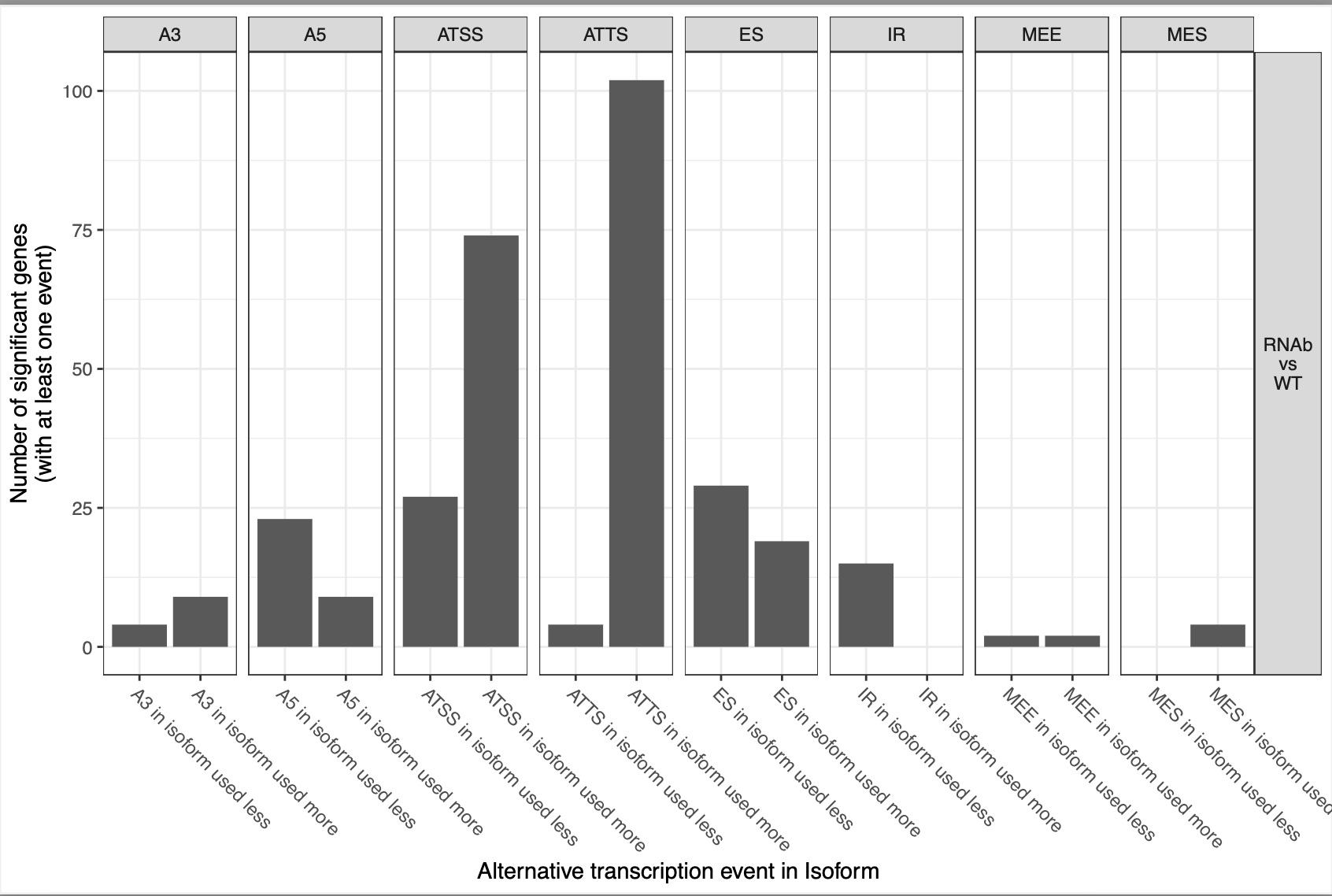
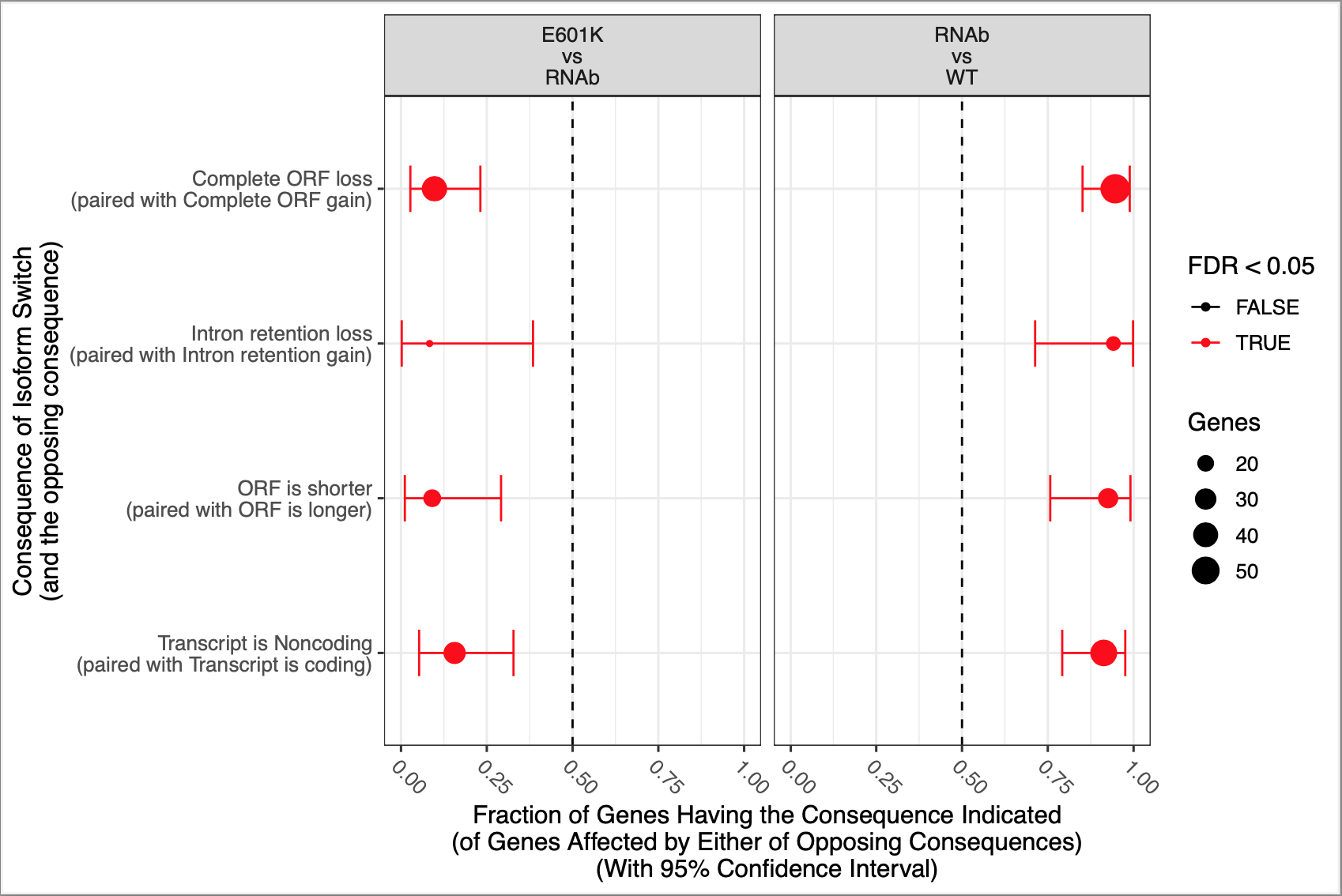
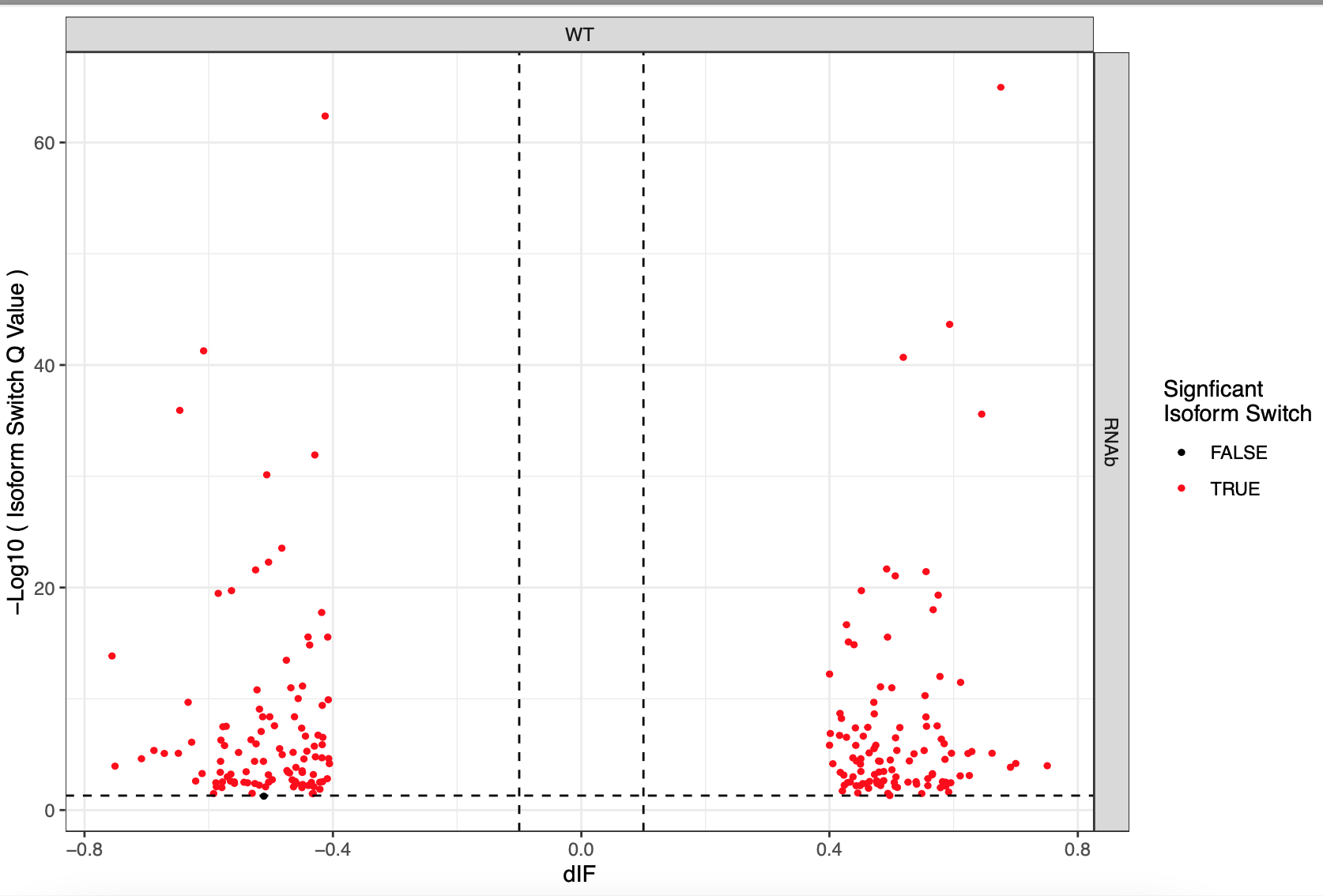
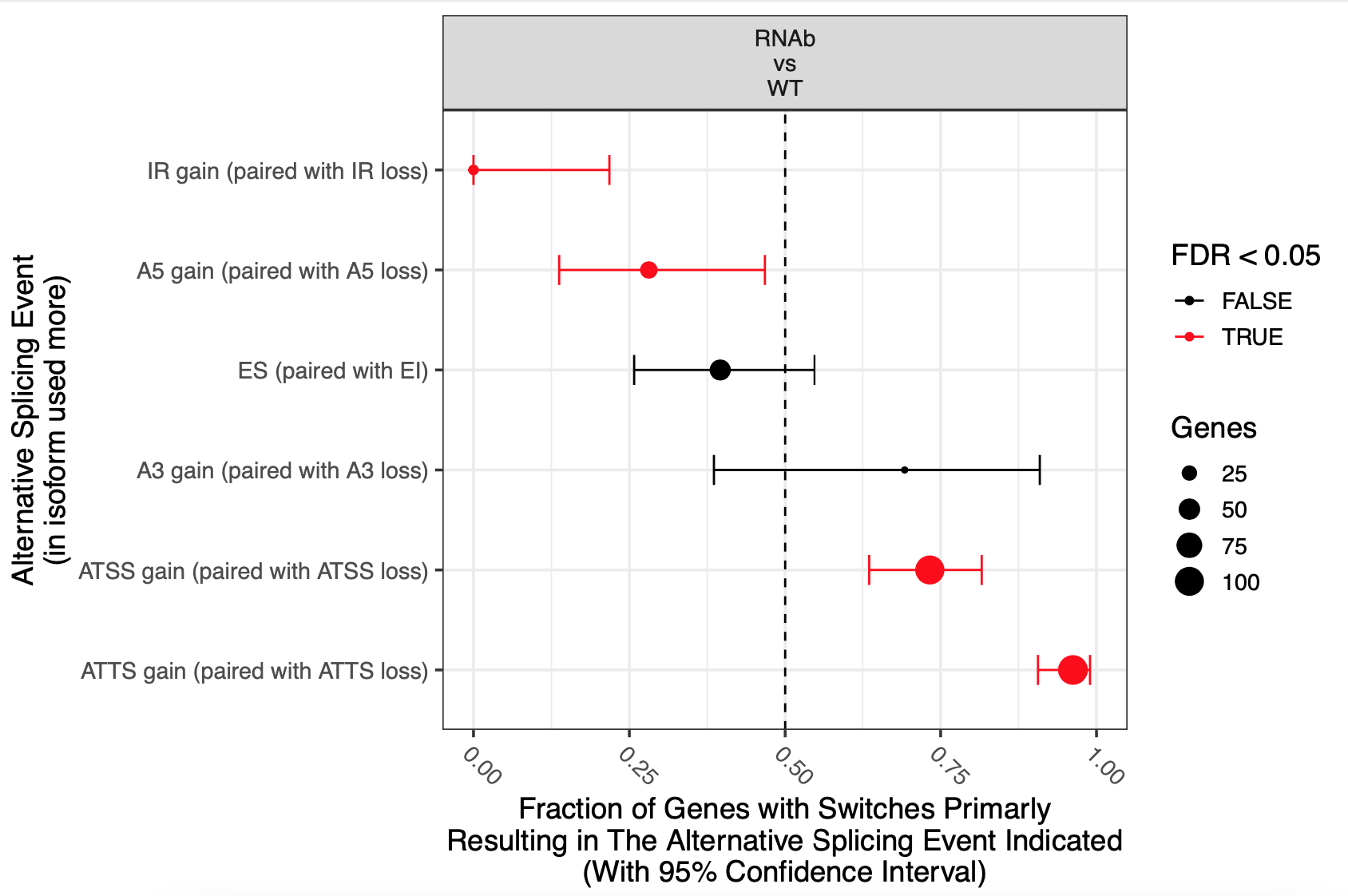
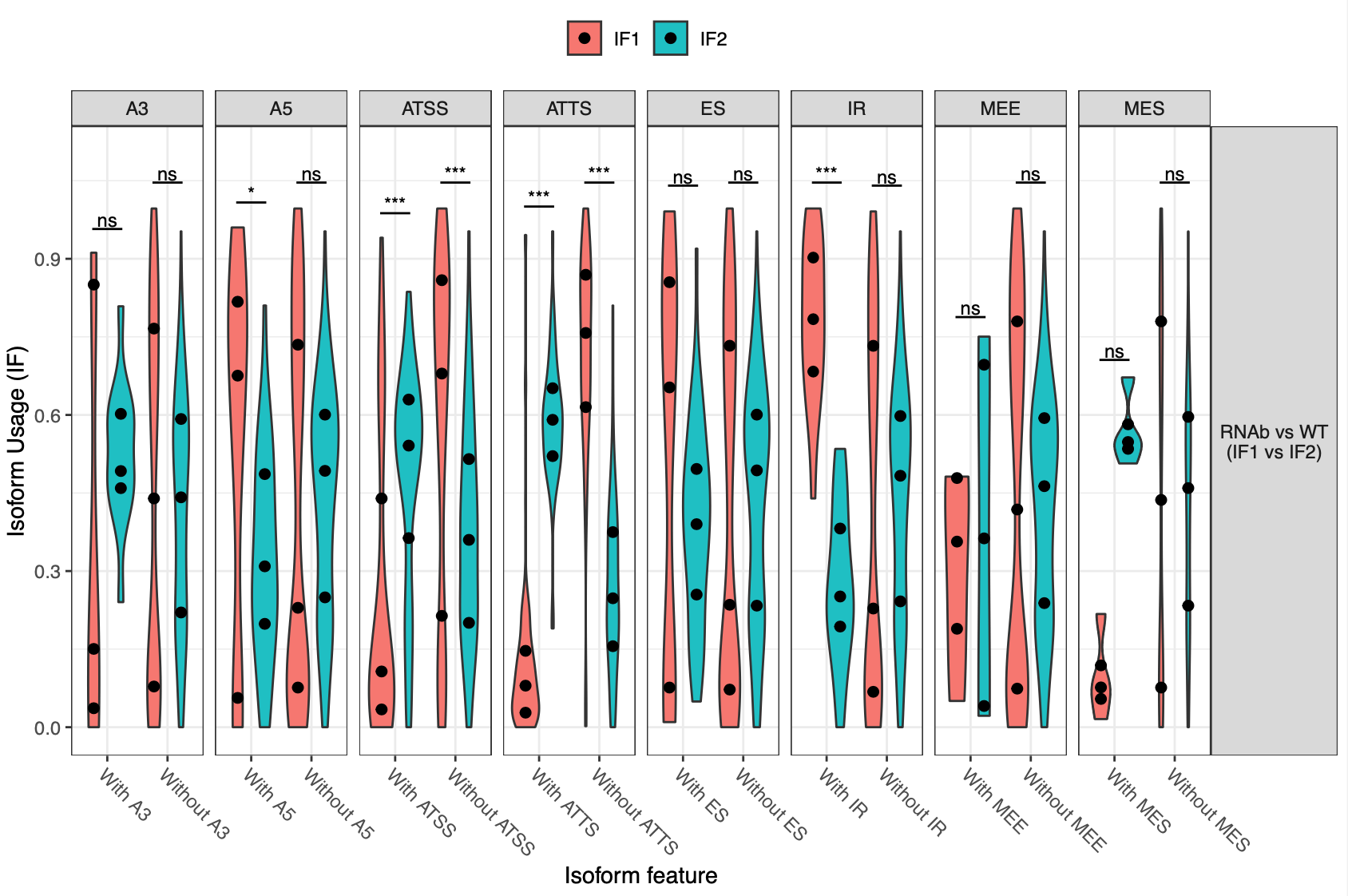
This is a snakemake pipeline to detect isoform switches from RNASeq data. The pipeline is in progress. It uses IsoformSwitchAnalyzer tool so far. The pipeline detects alternative splicing and differential isoform switchs. In addition, it relies on some external sources to analyze the consequences of those switches, including coding potential, ORF similarity, Intron retention, etc.
snakemake -jn
where n is the number of cores for example for 10 cores use:
snakemake -j10
For less froodiness, use conda:
snakemake -jn --use-conda
For example, for 10 cores use:
snakemake -j10 --use-conda
This will pull automatically the same versiosn of tools we used. Conda has to be installed in the system, in addition to snakemake.
For a dry run use:
snakemake -j1 -n
and to print command in dry run use:
snakemake -j1 -n -p
Just update your config file to include all your sample names, edit your interval.list file to include your intervals of interest, your path, etc for example:
snakemake -j1 --configfile config-WES.yaml
or:
snakemake -j1 configfile config-WGS.yaml
- Vitting-Seerup et al. The Landscape of Isoform Switches in Human Cancers. Cancer Res. (2017)