Inversion of multimode dispersion curves of surface wave
The forwarding program is from disba, which is a computationally efficient Python library for the modeling of surface wave dispersion that implements a subset of codes from Computer Programs in Seismology (CPS) in Python compiled just-in-time with numba.
The optimization part is based on python pacakge scipy.optimize,
where the gradient is implemented in C++[1], and converted into the library
that can be imported by python with the help of pybind11.
There are two circumstances: with sudo permission and without sudo permission. The prior is typically for PC, and the latter is usually for clusters.
-
Install git, eigen3 and cmake through the package manager according to the system, e.g. apt-get, homebrew, pacman.
-
Make sure your c++ compiler support c++14
-
Install python packages
- Install miniconda
> wget https://mirrors.tuna.tsinghua.edu.cn/anaconda/miniconda/Miniconda3-py38_4.9.2-Linux-x86_64.sh > bash ./Miniconda3-py38_4.9.2-Linux-x86_64.sh > source .bashrc
- Create environment
> conda create -n disba python=3.8 > conda activate disba > conda install numpy scipy matplotlib pyyaml mpi4py pandas tqdm > pip install disba mpi_master_slave
- Install miniconda
We use spack to install required softwares and libraries, which is a package manager for supercomputers, Linux, and macOS.
-
Install spack
> git clone https://github.com/spack/spack.git > echo ". spack/share/spack/setup-env.sh" >> .bashrc > source ~/.bashrc
-
Install gcc of higher version
> spack info gcc > spack install ${A_VERSION_OF_GCC} > spack location -i gcc # spack compiler find ${LOCATION_OF_GCC_NEW_VERSION} > spack compiler find $(spack location -i gcc)
${A_VERSION_OF_GCC} is a version of gcc which you can choose from the list that "spack info gcc" shows, e.g.
gcc@9.3.0.${LOCATION_OF_GCC_NEW_VERSION} is seen from "spack location -i gcc", e.g.
/home/test/spack/opt/spack/linux-centos7-haswell/gcc-4.8.5/gcc-9.3.0-swta24mlro2t4z7rfka55y3aum2ckm32 -
Install eigen3, git and cmake
> spack install eigen > spack install cmake > spack install git
-
Manage and load package using
lmod> spack install lmod > echo ". $(spack location -i lmod)/lmod/lmod/init/bash" >> .bashrc > source .bashrc > module avail
-
Load package and software
> module load ${MODULE_NAME}
${MODULE_NAME} can be found from "module avail", and we need to load gcc, cmake, git and eigen3. when I installed, the command was below:
> module load gcc-9.3.0-gcc-4.8.5-swta24m > module load cmake-3.18.4-gcc-9.3.0-nesdphq > module load git-2.29.0-gcc-9.3.0-y4b63yo > module load eigen-3.3.8-gcc-9.3.0-txefvoy
You can add these module load command to your .bashrc.
-
Install python package (the same procedure as above)
-
Install problems
- tar (child): lbzip2: cannot exec: no such file or directory
Solution:
> spack install bzip2 > export PATH=$(spack location -i bzip2):$PATH
- Error: COmmandNotFoundError: spack requires 'patch'
Solutiin:
> spack install patch > export PATH=$(spack location -i patch):$PATH
After installing requirements, the program is installed in the following.
> git clone https://github.com/pan3rock/DisbaTomo.git
> cd DisbaTomo
> git submodule init
> git submodule update
> mkdir build
> cd build
> cmake ..
> make -j4> cd .. # in {DisbaTomo/}
> mkdir examples
> cp -r demo/* examples/
> cd examples/regular-large
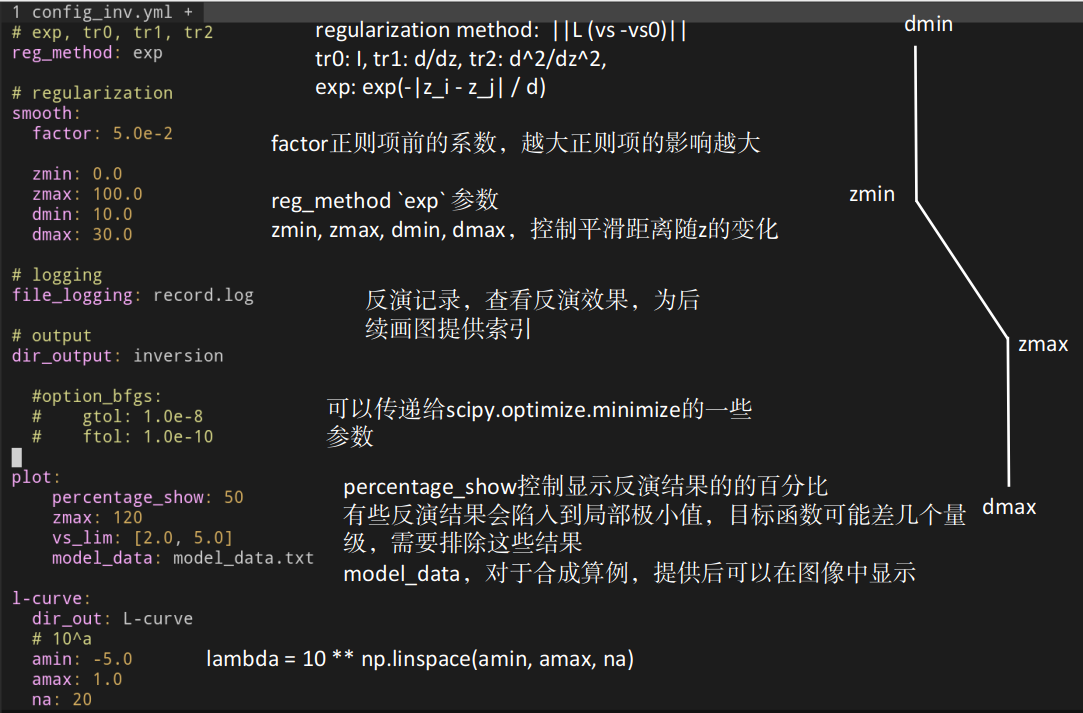
> mpirun -np 4 ../../python/determine_lcurve.py
> ../../python/plot_lcurve.py 0> mpirun -np 4 ../../python/inversion.py --num_init 20
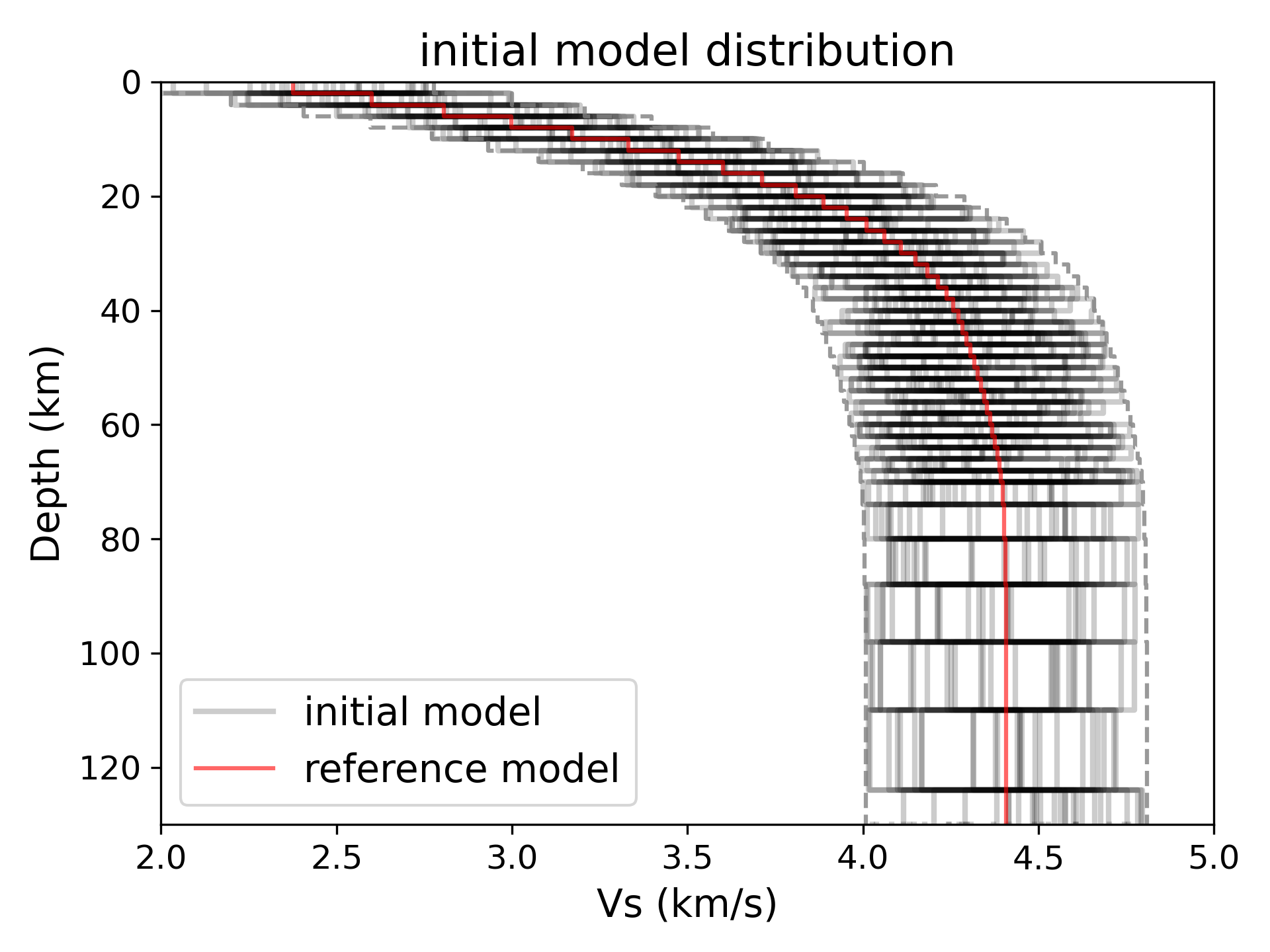
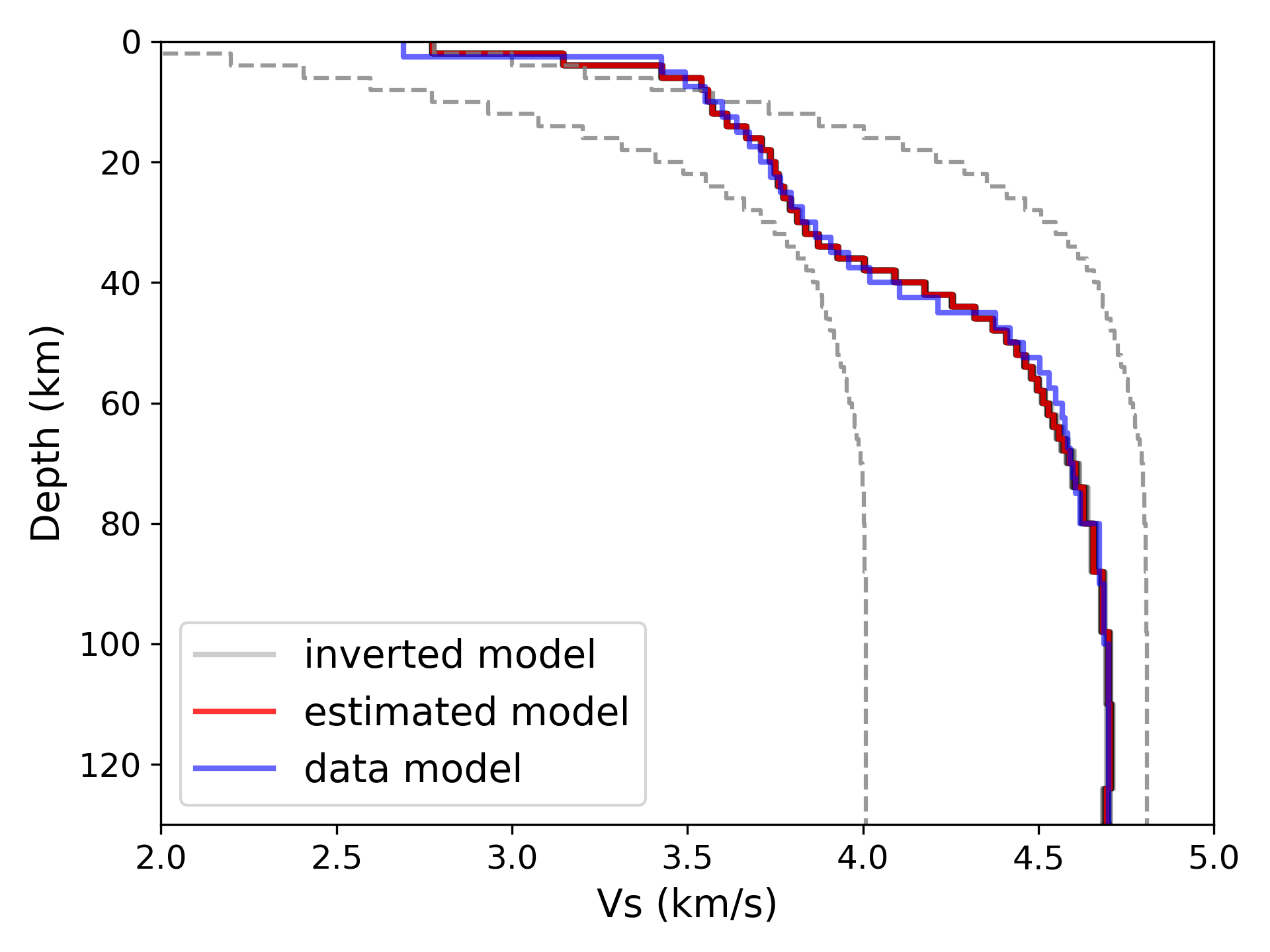
> ../../python/plot_inversion.py --data 0 --plot_model --plot_init
> ../../python/plot_inversion.py --data 0 --plot_model
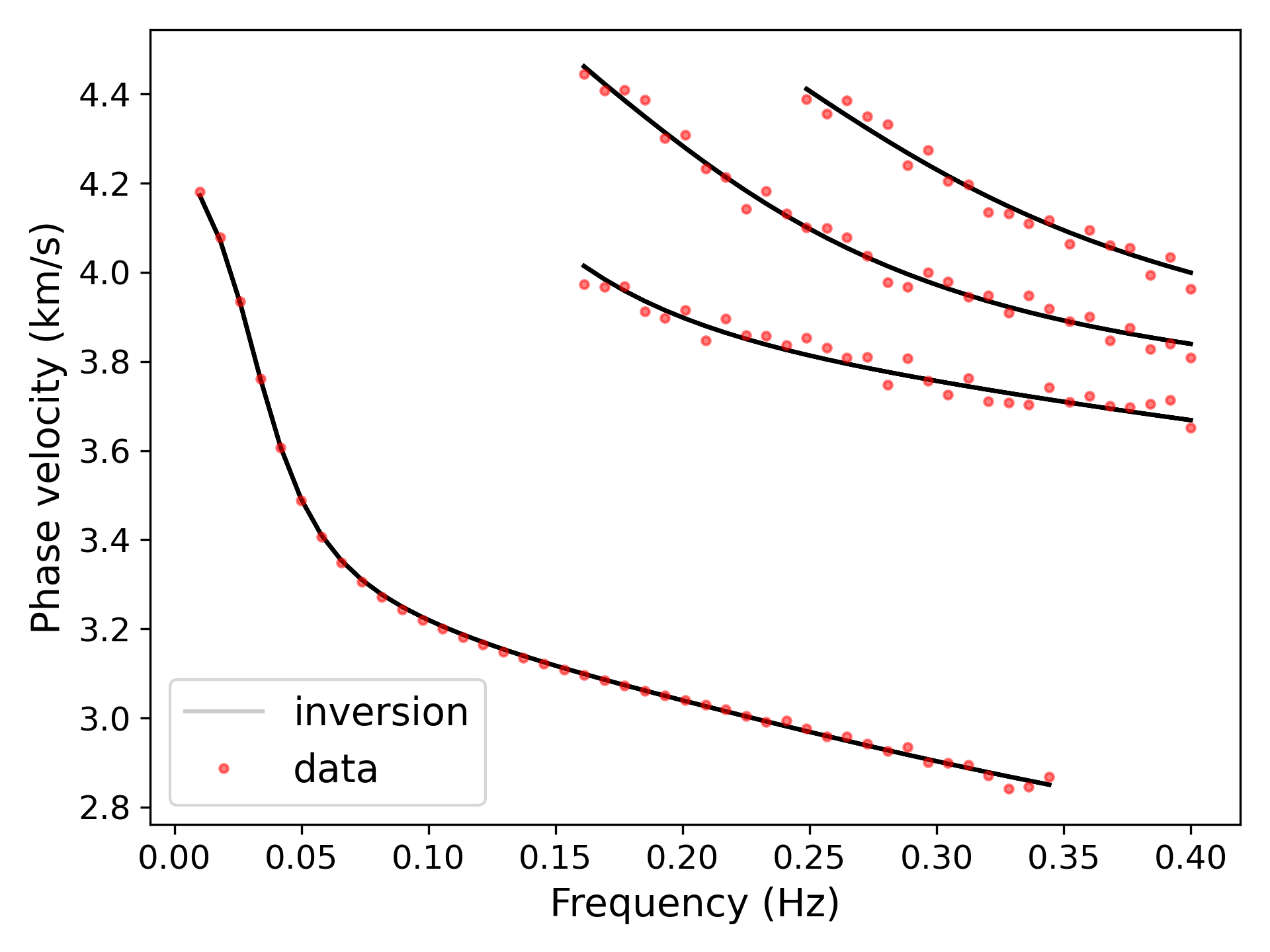
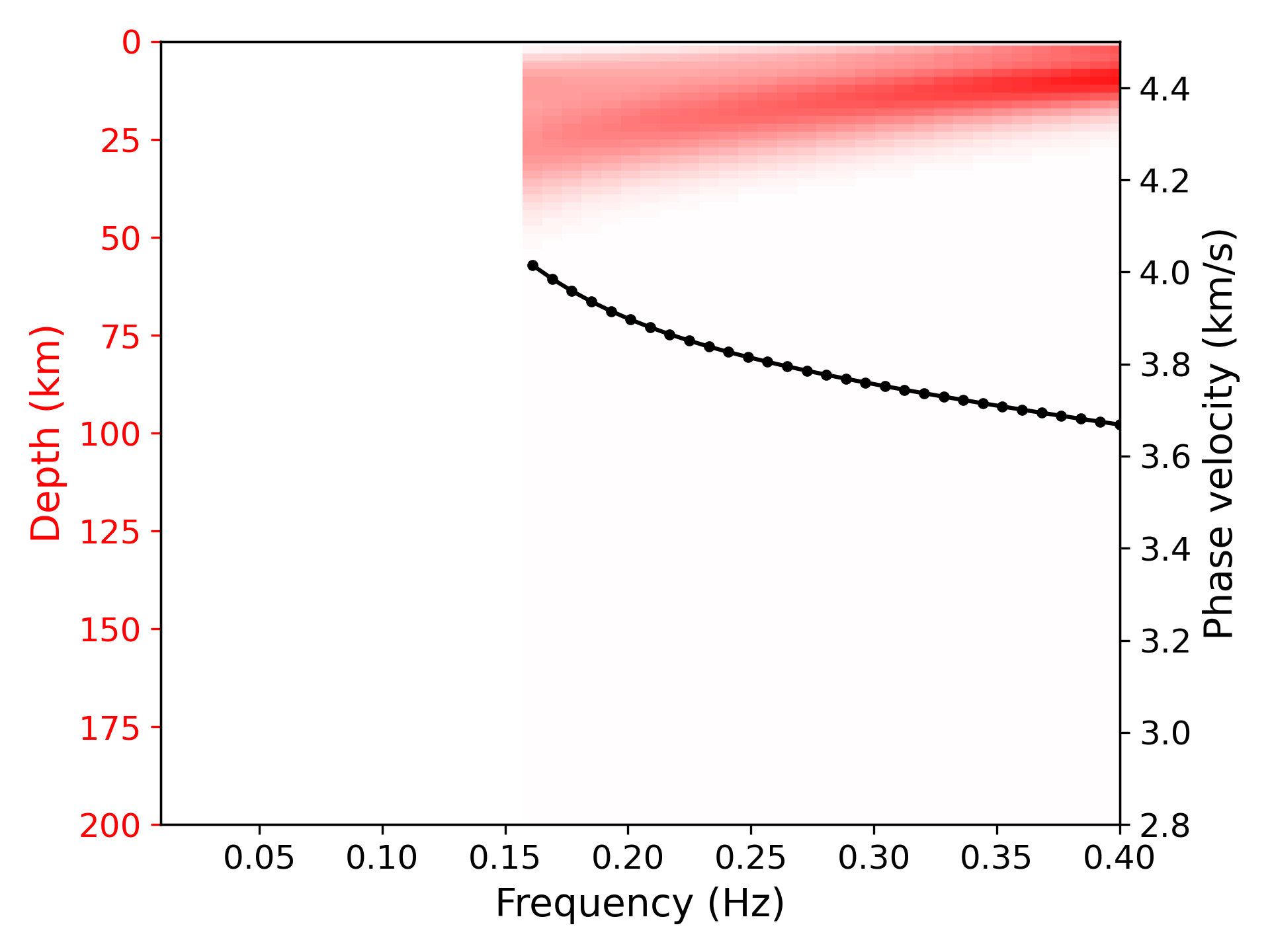
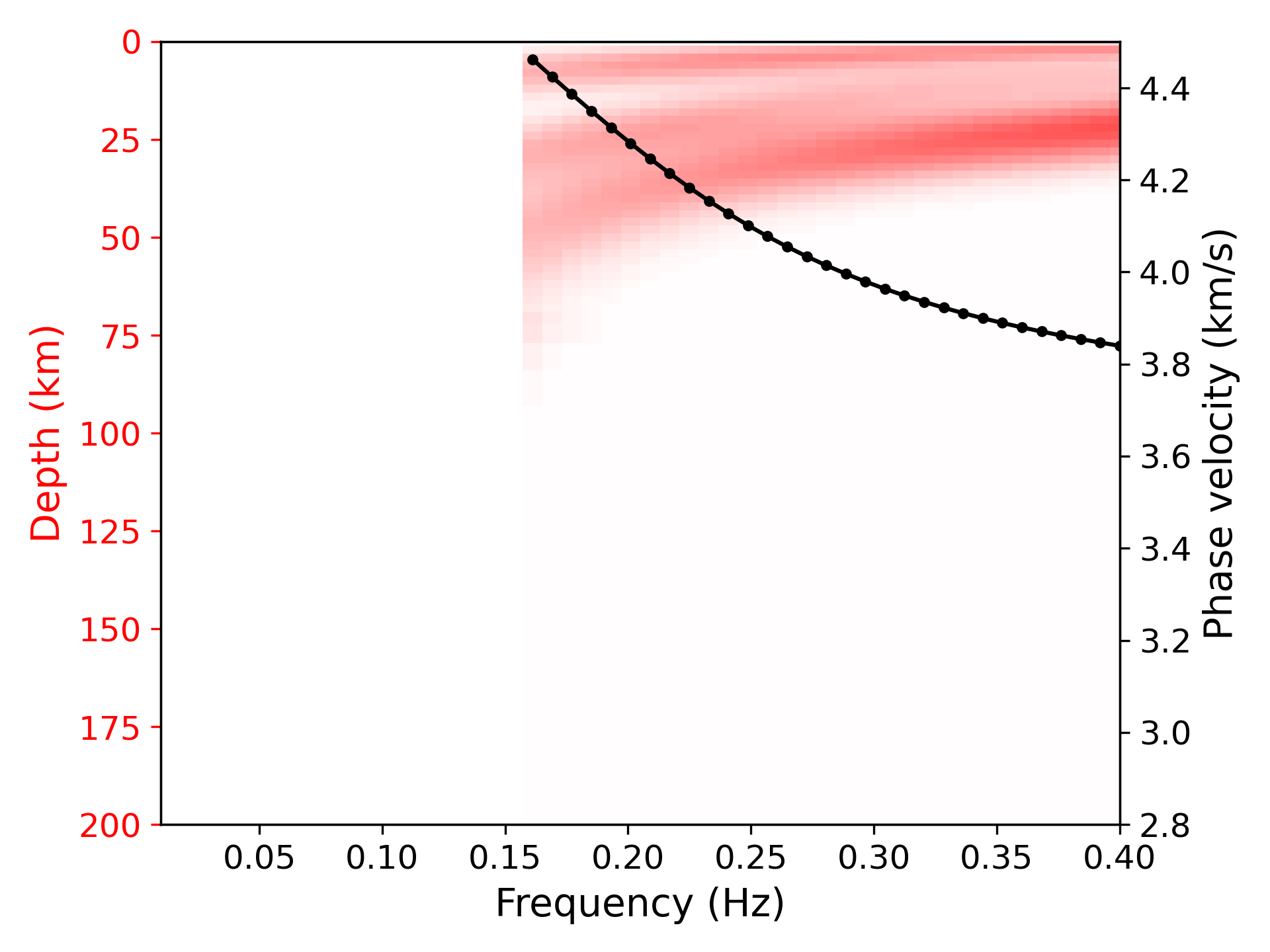
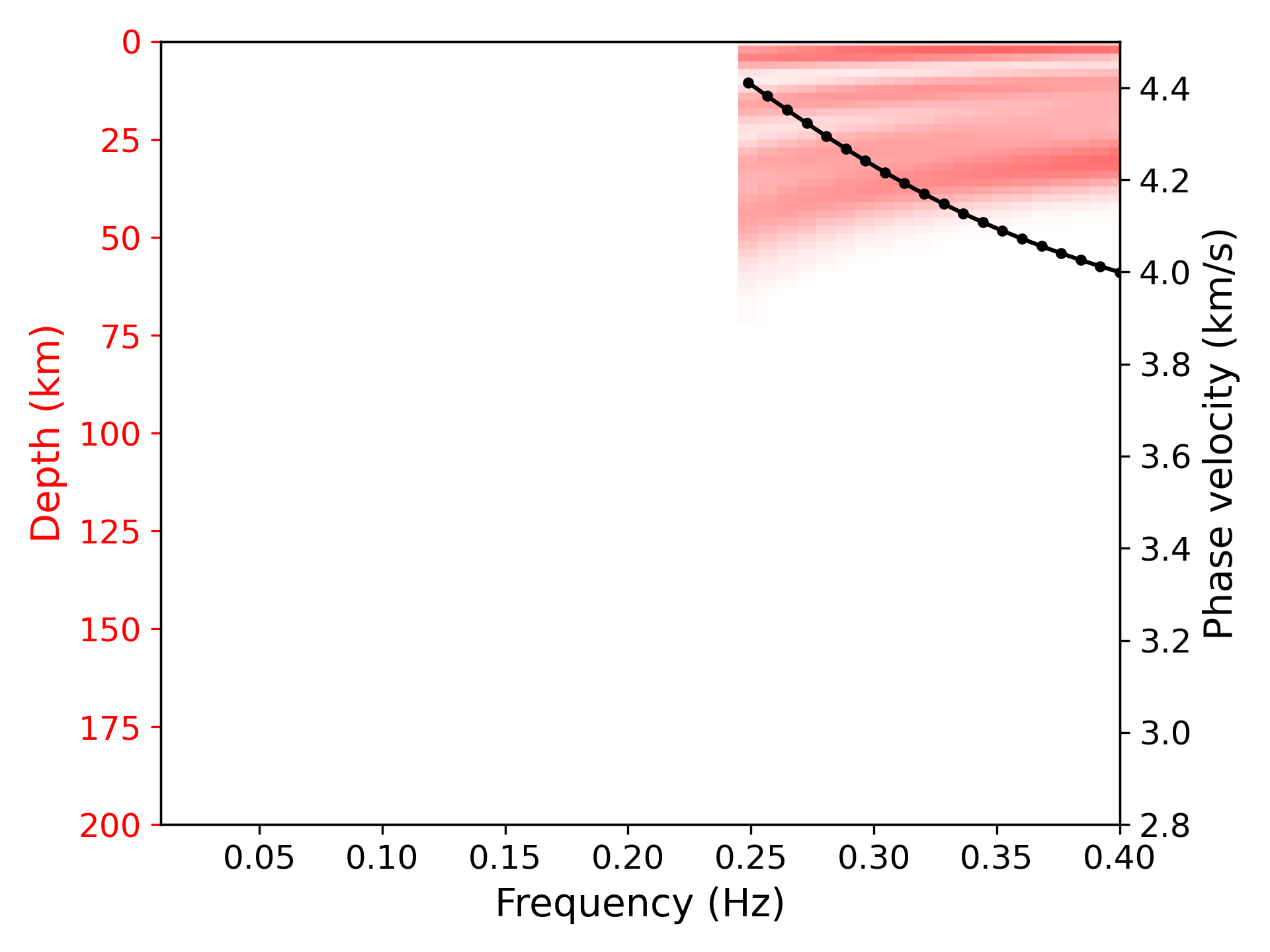
> ../../python/plot_inversion.py --data 0 --plot_dispThe sensitivity is defined in [1].
> ../../python/forward.py --data data/0.txt
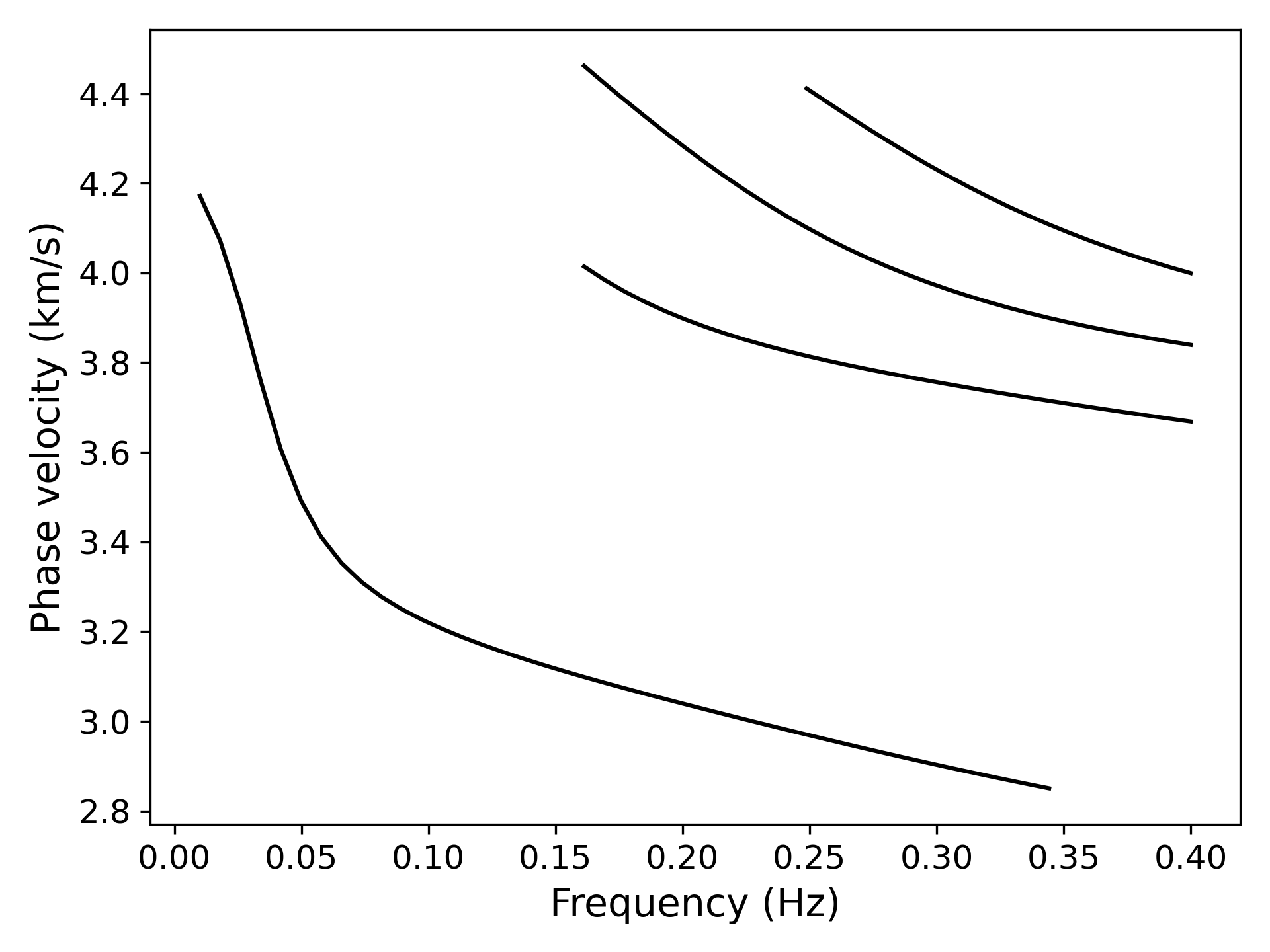
> ../../python/plot_disp.py disp.txt
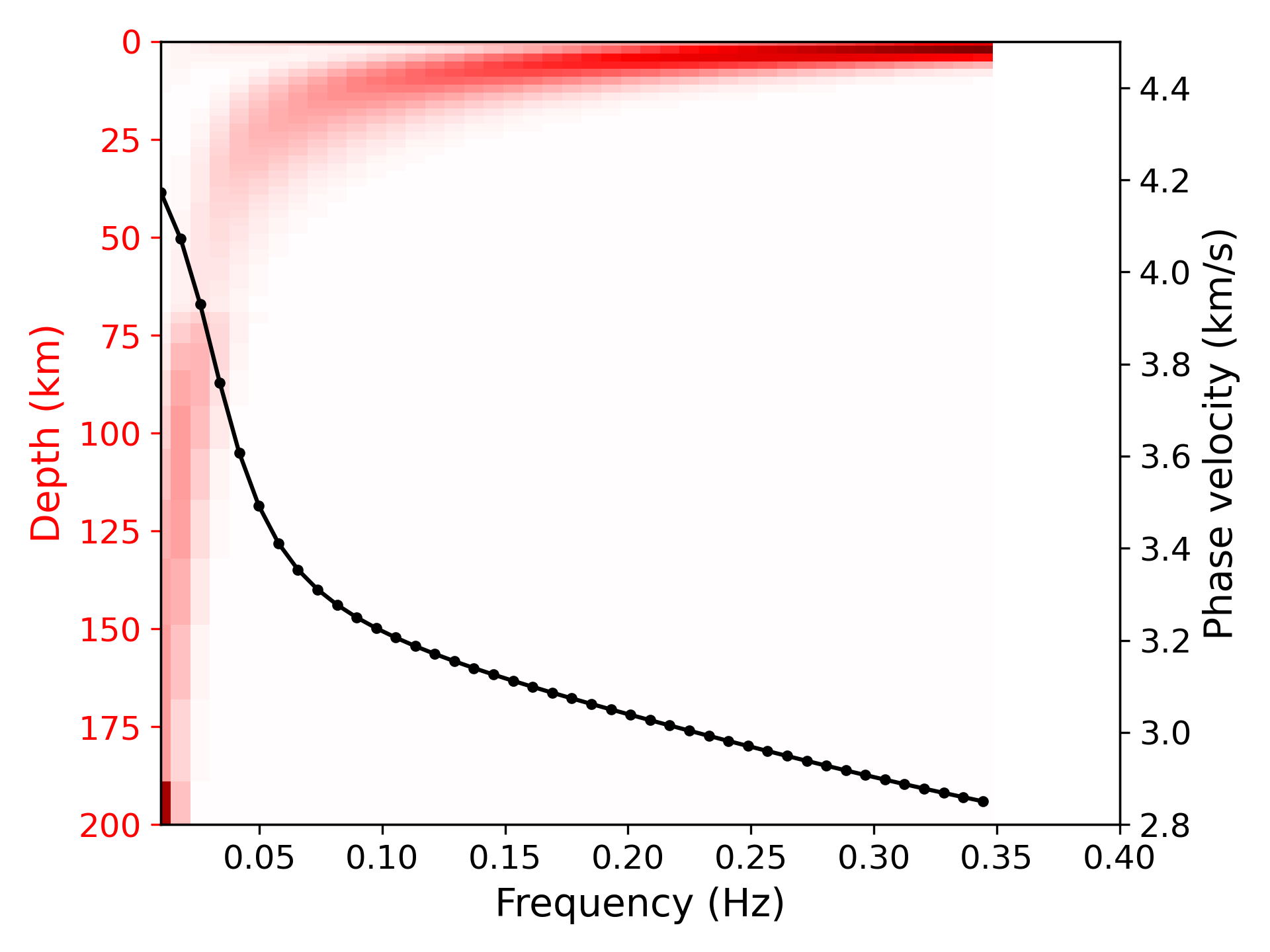
> ../../python/plot_kernel.py model_output.txt --disp disp.txt --fmin 0.01 --fmax 0.4 --cmin 2.8 --cmax 4.5 --plot_dispersion --vmax 0.35 --mode 0 --out k0.png
> ../../python/plot_kernel.py model_output.txt --disp disp.txt --fmin 0.01 --fmax 0.4 --cmin 2.8 --cmax 4.5 --plot_dispersion --vmax 0.35 --mode 1 --out k1.png
> ../../python/plot_kernel.py model_output.txt --disp disp.txt --fmin 0.01 --fmax 0.4 --cmin 2.8 --cmax 4.5 --plot_dispersion --vmax 0.35 --mode 2 --out k2.png
> ../../python/plot_kernel.py model_output.txt --disp disp.txt --fmin 0.01 --fmax 0.4 --cmin 2.8 --cmax 4.5 --plot_dispersion --vmax 0.35 --mode 3 --out k3.png> ../../python/forward.py -c config_forward2.yml --mode 3
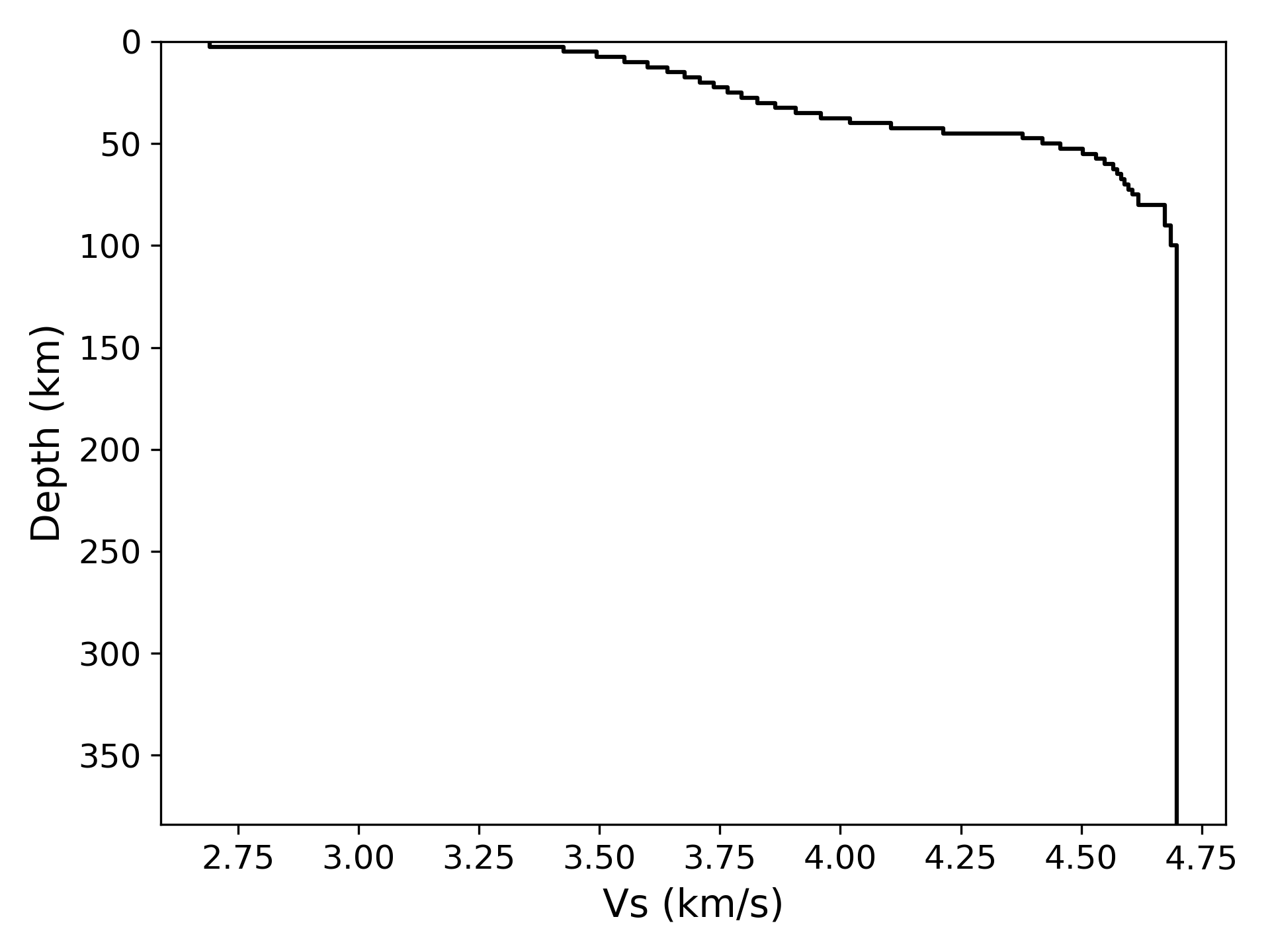
> ../../python/plot_model.py model_data.txt --out model_data.png
> ../../python/plot_disp.py disp.txt --out disp_data.png> ../../python/output_model.pyThe script will output estimated models to the directory model_inv, which name
is corresponding to the filename of data.
The estimated models are based on inverted models from different initial mdoesls
and their corresponding fitness value.
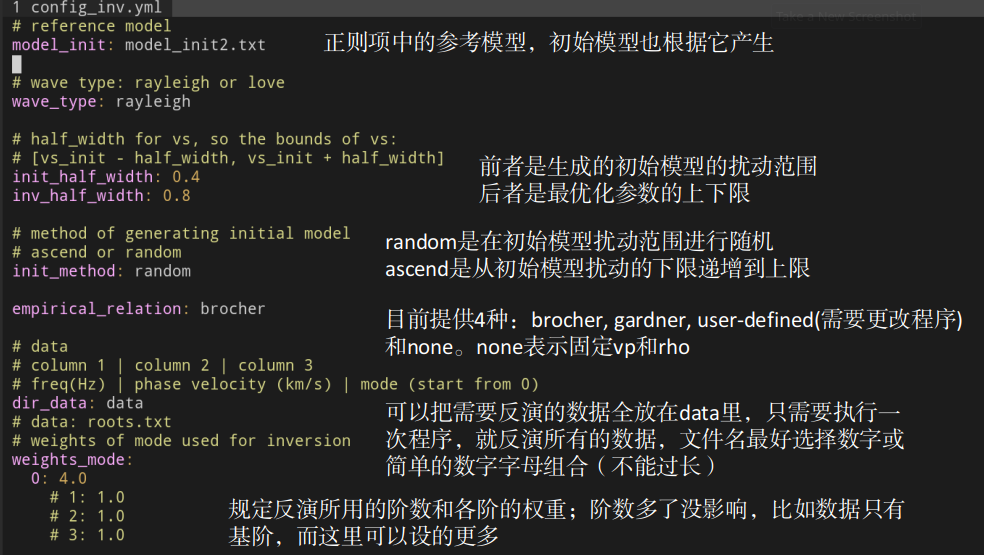
TODO: translate into English
Brocher and Gardner empirical relation have been provided, and you can add other
formula in python/empirical_relation.py.
[1] Pan, L., Chen, X., Wang, J., Yang, Z., & Zhang, D. (2019). Sensitivity analysis of dispersion curves of Rayleigh waves with fundamental and higher modes. Geophysical Journal International, 216(2), 1276–1303. https://doi.org/10.1093/gji/ggy479