- Introduction
- Features
- Requirements
- Installation
- Usage
- Project Structure
- Data
- Evaluation
- Explainable AI
- Timeline
- License
- Credits
Fault diagnosis is the process of diagnosis of fault in the machine after a failure occurs. To analyze the type of fault we use fault diagnosis. Here we used the Tennessee Eastman dataset. The Tennessee Eastman Process is a well-known benchmark process used to evaluate process control and fault detection methods. The goal of fault detection is to identify when the process is operating abnormally, which can lead to product quality issues, equipment damage, and safety hazards.
In this project, I trained ML models as mentioned below as well as a neural network model trained on Kggle GPU for the classification of the fault class type which has 52 features. I created a web dashboard which gives a glimpse of how prediction works If you are interested, you can try out. This predition also explains you which features are important for a particular feature with respect to model via explainable ai tools like Shap, Lime
For data analysis and model training refer
# Example installation steps
git clone [https://github.com/ShreyashChacharkar/Fault-diagnosis](https://github.com/ShreyashChacharkar/Fault-diagnosis)
pip install -r requirements.txtFor data analysis and model training refer
For running web based app
python web-dashboard/app.py
Project's directories and files. Highlight the purpose of important files or folders.
project-root/
|-- dataset/
| |-- raw/
| |-- processed/
|-- notebooks/
| |-- tep-fault-diagnosis-tree-classification.ipynb
|-- web-dashboard/
| |-- static/
| |-- style.css
| |-- script.js
| |-- templates/
| |-- index.html
| |-- layout.html
| |-- main.py
| |-- utlis.py
| |-- requirements.txt
|-- README.md
|-- requirements.txt
Dataset link: kaggle source
This dataverse contains the data referenced in Rieth et al. (2017). Issues and Advances in Anomaly Detection Evaluation for Joint Human-Automated Systems. To be presented at Applied Human Factors and Ergonomics 2017.
Each .RData file is an external representation of an R dataframe that can be read into an R environment with the 'load' function. The variables loaded are named ‘fault_free_training’, ‘fault_free_testing’, ‘faulty_testing’, and ‘faulty_training’, corresponding to the RData files.
Each dataframe contains 55 columns:
-
Column 1 ('faultNumber') ranges from 1 to 20 in the “Faulty” datasets and represents the fault type in the TEP. The “FaultFree” datasets only contain fault 0 (i.e. normal operating conditions).
-
Column 2 ('simulationRun') ranges from 1 to 500 and represents a different random number generator state from which a full TEP dataset was generated (Note: the actual seeds used to generate training and testing datasets were non-overlapping).
-
Column 3 ('sample') ranges either from 1 to 500 (“Training” datasets) or 1 to 960 (“Testing” datasets). The TEP variables (columns 4 to 55) were sampled every 3 minutes for a total duration of 25 hours and 48 hours respectively. Note that the faults were introduced 1 and 8 hours into the Faulty Training and Faulty Testing datasets, respectively.
-
Columns 4 to 55 contain the process variables; the column names retain the original variable names.
for data preprocessing refer:
#.rdata file reader
import pyreadr
#zipfile extract
import zipfile as zp
#extract from zipfile
with zp.ZipFile("dataset\TEP_FaultFree_Training.RData (1).zip", 'r') as zip_ref:
zip_ref.extractall("dataset")
# pyreadr converts rData into dictionary type object.
result = pyreadr.read_r("dataset/TEP_Faulty_Training.RData")
result1 = pyreadr.read_r("dataset/TEP_FaultFree_Training.RData")
#dataframes
df_train = result['faulty_training']
df_ff = result1['fault_free_training']
#save in csv file
df_ff.to_csv("dataset/fault_free_training.csv")
df_train.to_csv("dataset/faulty_training.csv")
| Method | Accuracy |
|---|---|
| XG Boost | 0.924 |
| Random Forest | 0.895 |
| Naive Bayes | 0.652 |
| KNN | 0.464 |
| Decision Tree | 0.827 |
| Logistic Regression | 0.695 |
| Neural Networks | 0.946 |
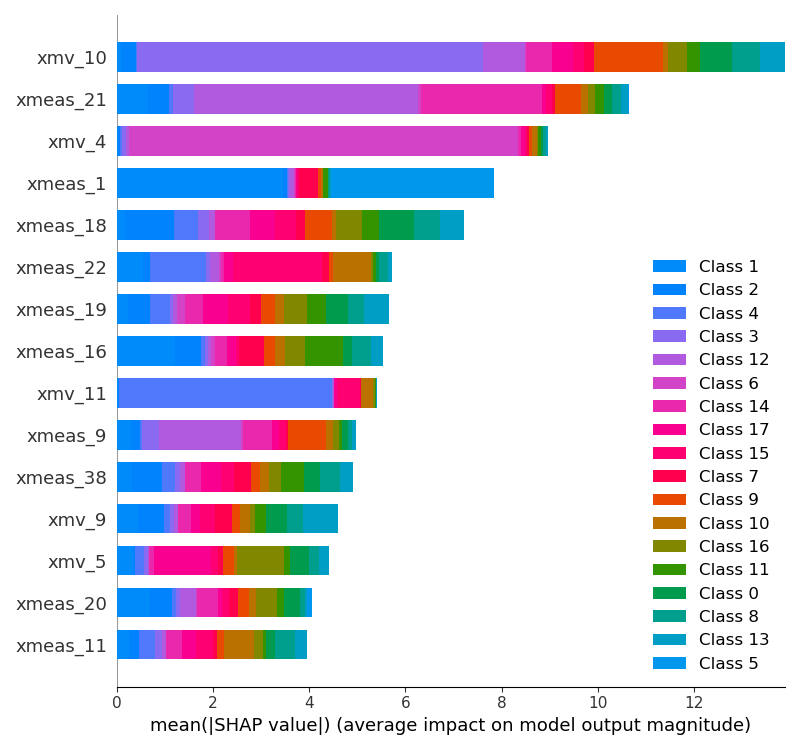
- Lime
- Permutation importance
- Partial dependence
- Shap (local)
- Shap (global)
eg.
| Days | Tasks | Description |
|---|---|---|
| Day 1 | Topic introduction | read research paper, youtube, article |
| Day 1 to 2 | Data wrangling | Data preprocessing (pyredr lib.), Cleaning, ETL activities, Data analysis, Data visualisation(matplotlib, seaborn) |
| Day 3 | Model Training | Training with data(sklearn, TensorFlow, classification algorithm), Feature extracting, Hyperparameter Tuning |
| Day 4 | Communication Result | Explainable AI Shaply and Lime, Real-time fault analysis |
| Day 5 to 8 | Web dashboard | Web dashboard (baseline with templates, style, app.py, utlis.py) |
| Day 9 | Deploying on cloud | Deploying selected ML model on GCP and AWS connect apis |
This GitHub repository can be used for educational purposes only