Last updated 2022-10-31
Water service boundaries (spatial polygons) delineate areas over which water is delivered from water systems to customers. Across the USA, some states (e.g., CA, TX, PA) maintain centralized water system boundary (henceforth, wsb) databases and make these accessible to the public. Publicly-accessible wsb data is not easily discoverable or cataloged for all states.
In this work, we build a reproducible pipeline to assimilate existing wsb data in the USA (labeled data). We then match water system names, cities served, and facility centroids with spatial boundaries for Census places. We also engineer features that predict the approximate spatial extent of these boundaries and train statistical and machine learning models on these features to produce proxy water system boundaries for states without centralized wsb data. The result is a dataset of all potential geographic boundaries or features associated with each water system. We then apply a hierarchical selection, assigning the highest integrity spatial boundary (labeled water service area shapefile being the highest integrity; modeled boundary being the lowest integrity) to each water system. The result is a provisional water system boundary layer for active, community water systems across the US.
While this repository allows you to reproduce the data yourself, you may be here because you just want the end-data. To access the water service boundary map and data, please visit the SimpleLab Hydroshare and check for the latest version of the data, available for download.
The main function of the project is an ETM (extract-transform-model) pipeline that chains a set of modular programs which can be flexibly modified over time to accommodate changes in input data and required output results. The main output in {WSB_OUTPUT_PATH} is a filesystem of various spatial formats (e.g., shp, geojson, csv, rds) that are uploaded to Hydroshare for distribution.
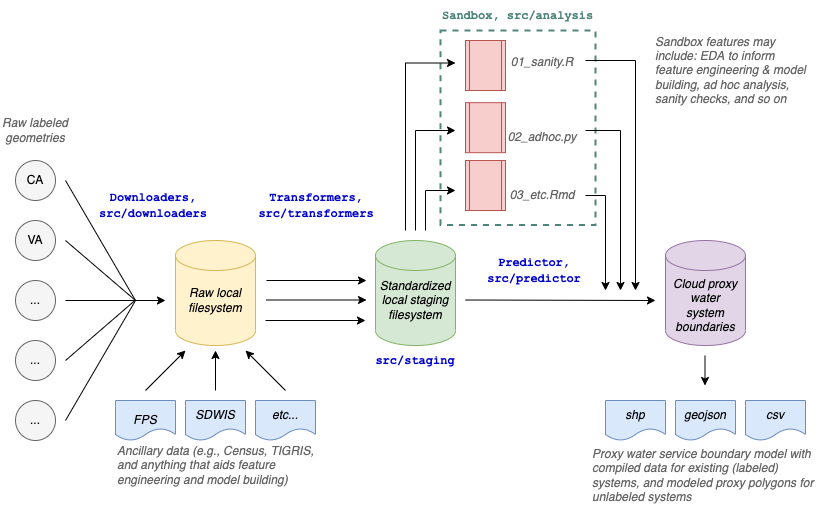
Download and transform data processing steps are modularized into separate processes in the /src/downloaders and src/transformers directories that can be modified and run in parallel, with no dependencies on one another. Downloaders pull raw data from the web to a local filesystem. See downloaders directory. Transformers clean, standardize and join that data, and then write it to data/staging. See transformers directory. The staged and standardized data is pulled into a matching model that joins all data sources together and assigns TIGER/Line places to water systems. See matching directory. Additionally, the transformed data is piped into a model, which combines the "best" centroids with a modeled radius to produce another map layer. See models directory. Finally, all layers are brought together and assigned tiers. See the combine_tiers step.
All data in this project is quite small and should easily fit into memory and on a PC.
Exploratory data analysis (EDA), sanity checks, and feature engineering experimentation occur in the sandbox (src/analysis) and are modularized into iterative notebooks and scripts that can serve multiple objectives without interfering with the functionality of the main ETM pipeline (i.e., src/run.py). This makes it easy to create and archive new analyses that serve a purpose, but that may never become productionized.
The data science contributor guide in the sandbox directory is a set of organizing guidelines for how EDA, analyses, and modeled output occur should be conducted in the sandbox.
The overall pipeline is shown below:
Clone this repo.
Install R version 4.2.1. Download packages as necessary. We rely only on version-stable CRAN packages.
Set environmental variables in two files: .env (python) and .Renviron (R):
From command line in your repositories folder you can:
touch .Renviron
touch .env
Note: The files .Renviron and .env need to be set up in the project root directory.
To add variables, open both environment files, copy and paste into each:
WSB_DATA_PATH = "path to save raw data from downloaders"
WSB_STAGING_PATH = "path to stage post-transformer data for EDA and modeling"
WSB_OUTPUT_PATH = "path to output matching artifacts, like reports"
WSB_EPSG = "4326"
WSB_EPSG_AW = "ESRI:102003"
POSTGIS_CONN_STR = "postgresql://postgres:postgres@localhost:5433/wsb"
CENSUS_API_KEY="<Get a census API key from https://api.census.gov/data/key_signup.html>"
Don't forget to leave a blank line at the end of .Renviron before saving.
To associate .Renviron with the R project, open R, run usethis::edit_r_environ(scope = "project").
WSB_DATA_PATH is where we save raw data from the downloaders, which may grow sizable.
WSB_STAGING_PATH is where we stage post-transformed for EDA and modeling.
POSTGIS_CONN_STR is the connection string for the local PostGIS docker database.
Use WSB_EPSG when writing to geopackages, and WSB_EPSG_AW when calculating areas on labeled geometries WSB_EPSG_AW is the coordinate reference system (CRS) used by transformers when we make calculations. We currently use Albers Equal Area Conic projected CRS for equal area calculations. For AK and HI, we need to shift geometry into this CRS so area calculations are minimally distorted, see tigris::shift_geometry(d, preserve_area = TRUE) at this webpage. WSB_EPSG is a World Geodetic System 1984 (see here) which is the CRS that geojson stores.
It's recommended to use a virtual environment of some sort. On Windows, you must use Conda, because Python venv has trouble with some of the packages in this repo.
You'll also need to install aria for downloading large files.
On Windows, the preferred method is to use the chocolatey package manager:
choco install aria2
On Mac:
brew install aria2
Self-solve system-specific issues.
We use a "snapshot and restore" approach to dependency management for Python and R environments. This may be superseded by Docker at a later time.
Download RStudio, then open wsb.RProject in RStudio. This will bootstrap renv. Next, in the R console, run renv::restore() to install R package dependencies and self-solve system-specific issues.
When a developer updates or installs new packages to the R project, the lockfile must be updated. Steps include:
- A user installs, or updates, one or more packages in their local project library;
- That user calls
renv::snapshot()to update therenv.locklockfile; - That user then shares the updated version of
renv.lockwith their collaborators by pushing to Github; - Other collaborators then call
renv::restore()to install the packages specified in the newly-updated lockfile.
To match and join all data including spatial data, you must have a PostGIS database. It's recommended to use a docker container. Follow the instructions as follows:
Download Docker for Mac or Windows, here.
Then, from your command line run:
docker run -e POSTGRES_PASSWORD=postgres -d --name postgis -p 5433:5432 -v postgis_volume:/var/lib/postgresql/data postgis/postgis
Some notes about this command:
- The database password will be "postgres". This is safe when run locally, but never use this on a server exposed to the internet.
- PostGIS will be available on port 5433
- The data will be stored in a named docker volume called
postgres_volume. This will preserve your data even if the container is stopped and removed.
Log into the container:
docker exec -it postgis bash
Connect to the PostGIS server using psql:
psql -U postgres
Create a new database:
create database wsb;
Connect to the database:
\c wsb
Add the PostGIS extension:
CREATE EXTENSION postgis;
Create the schema: Copy-paste the code from src/match/init_model.sql
Exit out of psql:
exit
Exit out of the docker container:
exit
You have now initialized the PostGIS database in a docker container and can run the pipeline.
To run the entire pipeline:
python run_pipeline.py
To run only a portion of the pipeline, open src/run_pipeline.py in an interactive Python IDE (such as Spyder or VSCode). Run the top cell to set up the session, then run the relevant cells.
Or, you can work with the specific scripts directly. Run all files in the following directories, following each repository's README, in this order:
src/downloaderssrc/transformerssrc/matchsrc/model
To contribute to the project, first read the contributing docs. Always branch from develop or a subbranch of develop and submit a pull request. To be considered as a maintainer, please contact Jess Goddard .