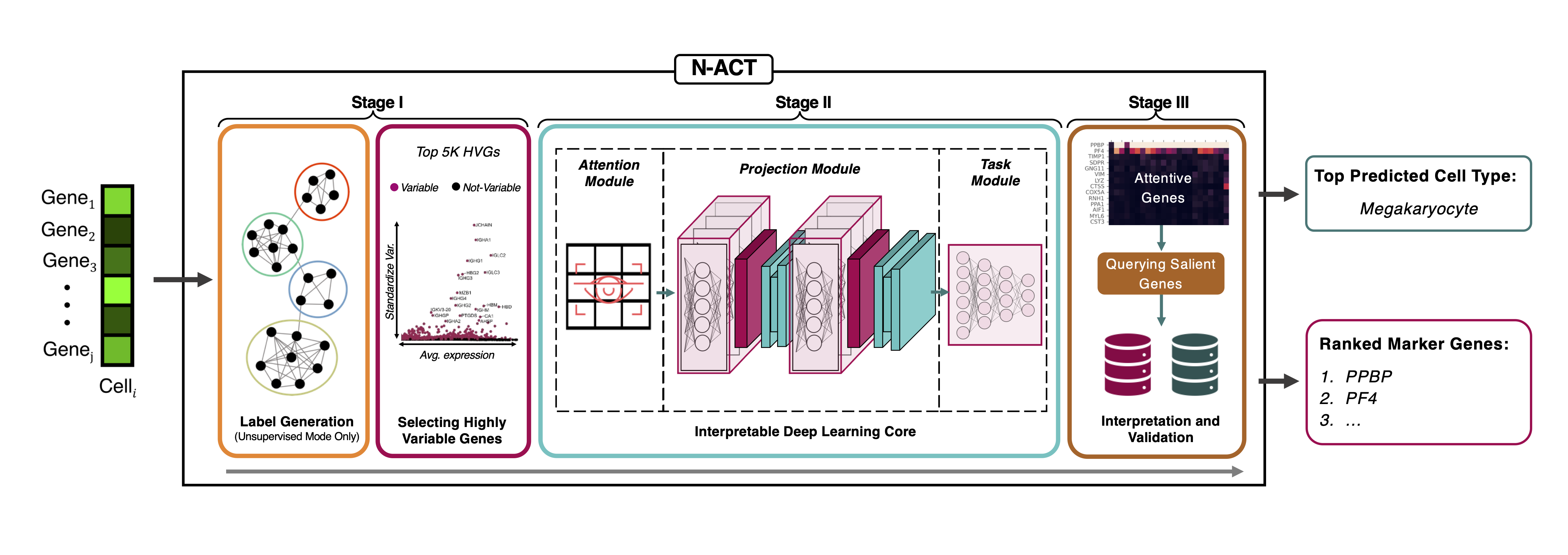
N-ACT (Package Repository)
This repository hosts the package for N-ACT: An Interpretable Deep Learning Model for Automatic Cell Type and Salient Gene Identification (WCB @ ICML2022 paper). To make package development and maintaining more efficient, we have located training scripts and tutorials in different repositories into different repositories, as listed below.
Installing N-ACT
Installing the GitHub Repository (Recommended)
N-ACT can be installed using PyPI:
$ pip install git+https://github.com/SindiLab/N-ACT.git
or can be first cloned and then installed as the following:
$ git clone https://github.com/SindiLab/N-ACT.git
$ pip install ./N-ACT
Install Package Locally with pip
Once the files are available, make sure to be in the same directory as setup.py. Then, using pip, run:
pip install -e .In the case that you want to install the requirements explicitly, you can do so by:
pip install -r requirements.txtAlthough the core requirements are listed directly in setup.py. Nonetheless, it is good to run this beforehand in case of any dependecies conflicts.
Training N-ACT
All main scripts for training our deep learning model are located in this separate repository.
Tutorials
We have compiled a set of notebooks as tutorials to showcase N-ACT's capabilities and interptretability. These notebooks located here.
Please feel free to open issues for any questions or requests for additional tutorials!
Trained Models
TODO: Will be released with the next preprint for N-ACT.
Citation
If you found our work useful for your ressearch, please cite our preprint:
@article {Heydari2022.05.12.491682,
author = {Heydari, A. Ali and Davalos, Oscar A. and Hoyer, Katrina K. and Sindi, Suzanne S.},
title = {N-ACT: An Interpretable Deep Learning Model for Automatic Cell Type and Salient Gene Identification},
elocation-id = {2022.05.12.491682},
year = {2022},
doi = {10.1101/2022.05.12.491682},
journal = {The 2022 International Conference on Machine Learning (ICML) Workshop on Computational Biology Proceedings.},
URL = {https://www.biorxiv.org/content/early/2022/05/13/2022.05.12.491682},
eprint = {https://www.biorxiv.org/content/early/2022/05/13/2022.05.12.491682.full.pdf},
}