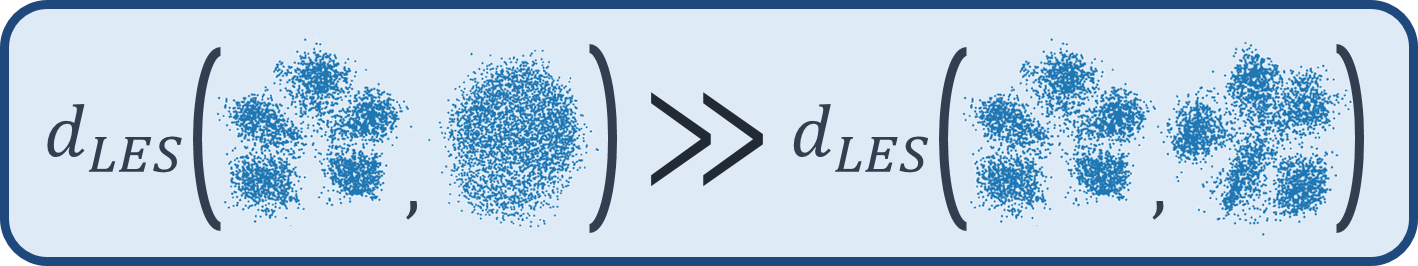Python implementation of the LES distance and examples from the paper. LES is an intrinsic geometric measure for comparing unaligned datasets of different dimensions and domains.
- Numpy
- SciPy
- Matplotlib
- Optional packages for clustered data usage example: scikit-learn
- Optional packages and repositories for comparisons with other algorithms:
The code was tested with python 3.8 and 3.9
import numpy as np
import sklearn.datasets as skld
from les.les import les_desc_comp, les_dist_comp
np.random.seed(1)
x1 = skld.make_blobs(n_samples=1000, centers=5, n_features=10)[0]
x2 = skld.make_blobs(n_samples=1000, centers=5, n_features=9)[0]
x3 = skld.make_blobs(n_samples=1000, centers=1, n_features=10)[0]
SIGMA, NEV, GAMMA = 2, 200, 1e-8
les_desc_x1 = les_desc_comp(x1, SIGMA, NEV, GAMMA)
les_desc_x2 = les_desc_comp(x2, SIGMA, NEV, GAMMA)
les_desc_x3 = les_desc_comp(x3, SIGMA, NEV, GAMMA)
print('LES(x1{5 clusters}, x2{5 clusters})=',les_dist_comp(les_desc_x1, les_desc_x2))
print('LES(x1{5 clusters}, x3{1 cluster})=',les_dist_comp(les_desc_x1, les_desc_x3))
print('LES(x2{5 clusters}, x3{1 cluster})=',les_dist_comp(les_desc_x2, les_desc_x3))python les/main_tori.py
By default the code compares LES only with our implementation of IMD (our approach) to reduce run-time and code
dependencies. To compare with additional algorithms that were mentioned in the paper, specify the algorithm names in main_tori.py, ALGS2COMPARE, and follow
the installation instructions of each package in the prequisites above.
Note that the topological data analysis methods (tda) and Gromov-Wasserstein distance (gw) require significantly longer
run-times.
[1] Tsitsulin et al., "The Shape of Data: Intrinsic Distance for Data Distributions", ICLR, 2019.
[2] Khrulkov and Oseledets, "Geometry score: A method for comparing generative adversarial networks", ICML, 2018.
[3] Peyré et al., "Gromov-Wasserstein averaging of kernel and distance matrices", ICML, 2016.