rgbif gives you access to data from GBIF via their REST API. GBIF versions their API - we are currently using v1 of their API. You can no longer use their old API in this package - see ?rgbif-defunct.
Tutorials:
- rgbif vignette - the intro to the package
- issues vignette - how to clean GBIF data
- taxonomic names - examples of some confusing bits
The rgbif package API follows the GBIF API, which has the following sections:
registry(http://www.gbif.org/developer/registry) - Metadata on datasets, and contributing organizations, installations, networks, and nodesrgbiffunctions:dataset_metrics(),dataset_search(),dataset_suggest(),datasets(),enumeration(),enumeration_country(),installations(),networks(),nodes(),organizations()- Registry also includes the GBIF OAI-PMH service, which includes GBIF registry
data only.
rgbiffunctions:gbif_oai_get_records(),gbif_oai_identify(),gbif_oai_list_identifiers(),gbif_oai_list_metadataformats(),gbif_oai_list_records(),gbif_oai_list_sets()
species(http://www.gbif.org/developer/species) - Species names and metadatargbiffunctions:name_backbone(),name_lookup(),name_suggest(),name_usage()
occurrences(http://www.gbif.org/developer/occurrence) - Occurrences, both for the search and download APIsrgbiffunctions:occ_count(),occ_data(),occ_download(),occ_download_cancel(),occ_download_cancel_staged(),occ_download_get(),occ_download_import(),occ_download_list(),occ_download_meta(),occ_get(),occ_issues(),occ_issues_lookup(),occ_metadata(),occ_search()
The GBIF maps API (http://www.gbif.org/developer/maps) is not implemented in rgbif,
and are meant more for intergration with web based maps.
install.packages("rgbif")Alternatively, install development version
install.packages("devtools")
devtools::install_github("ropensci/rgbif")library("rgbif")Note: Windows users have to first install Rtools to use devtools
occ_search(scientificName = "Ursus americanus", limit = 50)
#> Records found [8424]
#> Records returned [50]
#> No. unique hierarchies [1]
#> No. media records [43]
#> No. facets [0]
#> Args [scientificName=Ursus americanus, limit=50, offset=0, fields=all]
#> # A tibble: 50 × 68
#> name key decimalLatitude decimalLongitude
#> <chr> <int> <dbl> <dbl>
#> 1 Ursus americanus 1229610234 44.06062 -71.92692
#> 2 Ursus americanus 1253300445 44.65481 -72.67270
#> 3 Ursus americanus 1229610216 44.06086 -71.92712
#> 4 Ursus americanus 1249277297 35.76789 -75.80894
#> 5 Ursus americanus 1249296297 39.08590 -105.24586
#> 6 Ursus americanus 1253314877 49.25782 -122.82786
#> 7 Ursus americanus 1249284297 43.68723 -72.32891
#> 8 Ursus americanus 1272078411 44.41793 -72.70709
#> 9 Ursus americanus 1262389246 43.80871 -72.20964
#> 10 Ursus americanus 1257415362 44.32746 -72.41007
#> # ... with 40 more rows, and 64 more variables: issues <chr>,
#> # datasetKey <chr>, publishingOrgKey <chr>, publishingCountry <chr>,
#> # protocol <chr>, lastCrawled <chr>, lastParsed <chr>, crawlId <int>,
#> # extensions <chr>, basisOfRecord <chr>, taxonKey <int>,
#> # kingdomKey <int>, phylumKey <int>, classKey <int>, orderKey <int>,
#> # familyKey <int>, genusKey <int>, speciesKey <int>,
#> # scientificName <chr>, kingdom <chr>, phylum <chr>, order <chr>,
#> # family <chr>, genus <chr>, species <chr>, genericName <chr>,
#> # specificEpithet <chr>, taxonRank <chr>, dateIdentified <chr>,
#> # year <int>, month <int>, day <int>, eventDate <chr>, modified <chr>,
#> # lastInterpreted <chr>, references <chr>, license <chr>,
#> # identifiers <chr>, facts <chr>, relations <chr>, geodeticDatum <chr>,
#> # class <chr>, countryCode <chr>, country <chr>, rightsHolder <chr>,
#> # identifier <chr>, verbatimEventDate <chr>, datasetName <chr>,
#> # verbatimLocality <chr>, gbifID <chr>, collectionCode <chr>,
#> # occurrenceID <chr>, taxonID <chr>, recordedBy <chr>,
#> # catalogNumber <chr>, http...unknown.org.occurrenceDetails <chr>,
#> # institutionCode <chr>, rights <chr>, eventTime <chr>,
#> # identificationID <chr>, occurrenceRemarks <chr>,
#> # infraspecificEpithet <chr>, coordinateUncertaintyInMeters <dbl>,
#> # informationWithheld <chr>Or you can get the taxon key first with name_backbone(). Here, we select to only return the occurrence data.
key <- name_backbone(name='Helianthus annuus', kingdom='plants')$speciesKey
occ_search(taxonKey=key, limit=20)
#> Records found [21970]
#> Records returned [20]
#> No. unique hierarchies [1]
#> No. media records [15]
#> No. facets [0]
#> Args [taxonKey=3119195, limit=20, offset=0, fields=all]
#> # A tibble: 20 × 67
#> name key decimalLatitude decimalLongitude
#> <chr> <int> <dbl> <dbl>
#> 1 Helianthus annuus 1249279611 34.04810 -117.79884
#> 2 Helianthus annuus 1315048347 34.04377 -116.94136
#> 3 Helianthus annuus 1305118889 18.40386 -66.04487
#> 4 Helianthus annuus 1249286909 32.58747 -97.10081
#> 5 Helianthus annuus 1253308332 29.67463 -95.44804
#> 6 Helianthus annuus 1262375813 29.82586 -95.45604
#> 7 Helianthus annuus 1262385911 32.78328 -96.70352
#> 8 Helianthus annuus 1265544678 32.58747 -97.10081
#> 9 Helianthus annuus 1262379231 34.04911 -117.80066
#> 10 Helianthus annuus 1265560496 34.12861 -118.20700
#> 11 Helianthus annuus 1269541227 NA NA
#> 12 Helianthus annuus 1265895094 42.87784 -112.43226
#> 13 Helianthus annuus 1272087563 28.51021 -96.81979
#> 14 Helianthus annuus 1265590525 29.86693 -95.64667
#> 15 Helianthus annuus 1270045172 33.92958 -117.37322
#> 16 Helianthus annuus 1265553900 34.12932 -118.20648
#> 17 Helianthus annuus 1269543851 29.50991 -94.50006
#> 18 Helianthus annuus 1305119137 11.86735 -83.93555
#> 19 Helianthus annuus 1265590989 34.19005 -117.31644
#> 20 Helianthus annuus 1315048128 34.03212 -117.47091
#> # ... with 63 more variables: issues <chr>, datasetKey <chr>,
#> # publishingOrgKey <chr>, publishingCountry <chr>, protocol <chr>,
#> # lastCrawled <chr>, lastParsed <chr>, crawlId <int>, extensions <chr>,
#> # basisOfRecord <chr>, taxonKey <int>, kingdomKey <int>,
#> # phylumKey <int>, classKey <int>, orderKey <int>, familyKey <int>,
#> # genusKey <int>, speciesKey <int>, scientificName <chr>, kingdom <chr>,
#> # phylum <chr>, order <chr>, family <chr>, genus <chr>, species <chr>,
#> # genericName <chr>, specificEpithet <chr>, taxonRank <chr>,
#> # dateIdentified <chr>, year <int>, month <int>, day <int>,
#> # eventDate <chr>, modified <chr>, lastInterpreted <chr>,
#> # references <chr>, license <chr>, identifiers <chr>, facts <chr>,
#> # relations <chr>, geodeticDatum <chr>, class <chr>, countryCode <chr>,
#> # country <chr>, rightsHolder <chr>, identifier <chr>,
#> # verbatimEventDate <chr>, datasetName <chr>, verbatimLocality <chr>,
#> # gbifID <chr>, collectionCode <chr>, occurrenceID <chr>, taxonID <chr>,
#> # recordedBy <chr>, catalogNumber <chr>,
#> # http...unknown.org.occurrenceDetails <chr>, institutionCode <chr>,
#> # rights <chr>, eventTime <chr>, identificationID <chr>,
#> # coordinateUncertaintyInMeters <dbl>, occurrenceRemarks <chr>,
#> # informationWithheld <chr>Get the keys first with name_backbone(), then pass to occ_search()
splist <- c('Accipiter erythronemius', 'Junco hyemalis', 'Aix sponsa')
keys <- sapply(splist, function(x) name_backbone(name=x)$speciesKey, USE.NAMES=FALSE)
occ_search(taxonKey=keys, limit=5, hasCoordinate=TRUE)
#> Occ. found [2480598 (22), 2492010 (3042553), 2498387 (971214)]
#> Occ. returned [2480598 (5), 2492010 (5), 2498387 (5)]
#> No. unique hierarchies [2480598 (1), 2492010 (1), 2498387 (1)]
#> No. media records [2480598 (1), 2492010 (5), 2498387 (1)]
#> No. facets []
#> Args [taxonKey=2480598,2492010,2498387, hasCoordinate=TRUE, limit=5,
#> offset=0, fields=all]
#> First 10 rows of data from 2480598
#>
#> # A tibble: 5 × 82
#> name key decimalLatitude decimalLongitude
#> <chr> <int> <dbl> <dbl>
#> 1 Accipiter erythronemius 920169861 -20.55244 -56.64104
#> 2 Accipiter erythronemius 920184036 -20.76029 -56.71314
#> 3 Accipiter erythronemius 1001096527 -27.58000 -58.66000
#> 4 Accipiter erythronemius 1001096518 -27.92000 -59.14000
#> 5 Accipiter erythronemius 686297260 5.26667 -60.73333
#> # ... with 78 more variables: issues <chr>, datasetKey <chr>,
#> # publishingOrgKey <chr>, publishingCountry <chr>, protocol <chr>,
#> # lastCrawled <chr>, lastParsed <chr>, crawlId <int>, extensions <chr>,
#> # basisOfRecord <chr>, taxonKey <int>, kingdomKey <int>,
#> # phylumKey <int>, classKey <int>, orderKey <int>, familyKey <int>,
#> # genusKey <int>, speciesKey <int>, scientificName <chr>, kingdom <chr>,
#> # phylum <chr>, order <chr>, family <chr>, genus <chr>, species <chr>,
#> # genericName <chr>, specificEpithet <chr>, taxonRank <chr>,
#> # coordinateUncertaintyInMeters <dbl>, year <int>, month <int>,
#> # day <int>, eventDate <chr>, lastInterpreted <chr>, license <chr>,
#> # identifiers <chr>, facts <chr>, relations <chr>, geodeticDatum <chr>,
#> # class <chr>, countryCode <chr>, country <chr>, recordedBy <chr>,
#> # catalogNumber <chr>, institutionCode <chr>, locality <chr>,
#> # gbifID <chr>, collectionCode <chr>, modified <chr>, identifier <chr>,
#> # created <chr>, occurrenceID <chr>, associatedSequences <chr>,
#> # higherClassification <chr>, taxonID <chr>, sex <chr>,
#> # establishmentMeans <chr>, continent <chr>, references <chr>,
#> # institutionID <chr>, dynamicProperties <chr>, fieldNumber <chr>,
#> # language <chr>, type <chr>, preparations <chr>,
#> # occurrenceStatus <chr>, rights <chr>, verbatimEventDate <chr>,
#> # higherGeography <chr>, nomenclaturalCode <chr>, endDayOfYear <chr>,
#> # georeferenceVerificationStatus <chr>, datasetName <chr>,
#> # verbatimLocality <chr>, otherCatalogNumbers <chr>,
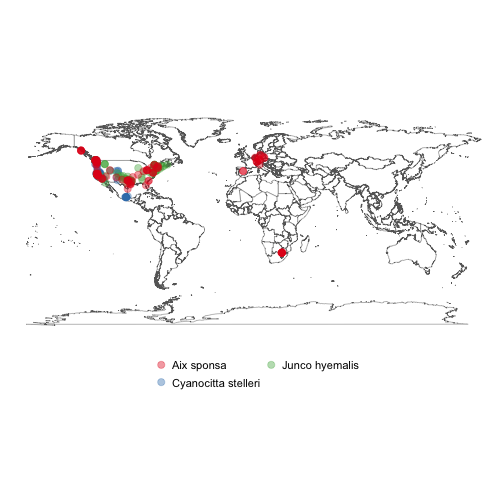
#> # startDayOfYear <chr>, accessRights <chr>, collectionID <chr>Make a simple map of species occurrences.
splist <- c('Cyanocitta stelleri', 'Junco hyemalis', 'Aix sponsa')
keys <- sapply(splist, function(x) name_backbone(name=x)$speciesKey, USE.NAMES=FALSE)
dat <- occ_search(taxonKey=keys, limit=100, return='data', hasCoordinate=TRUE)
library('plyr')
datdf <- ldply(dat)
gbifmap(datdf)- Please report any issues or bugs.
- License: MIT
- Get citation information for
rgbifin R doingcitation(package = 'rgbif') - Please note that this project is released with a Contributor Code of Conduct. By participating in this project you agree to abide by its terms.
This package is part of a richer suite called spocc - Species Occurrence Data, along with several other packages, that provide access to occurrence records from multiple databases.