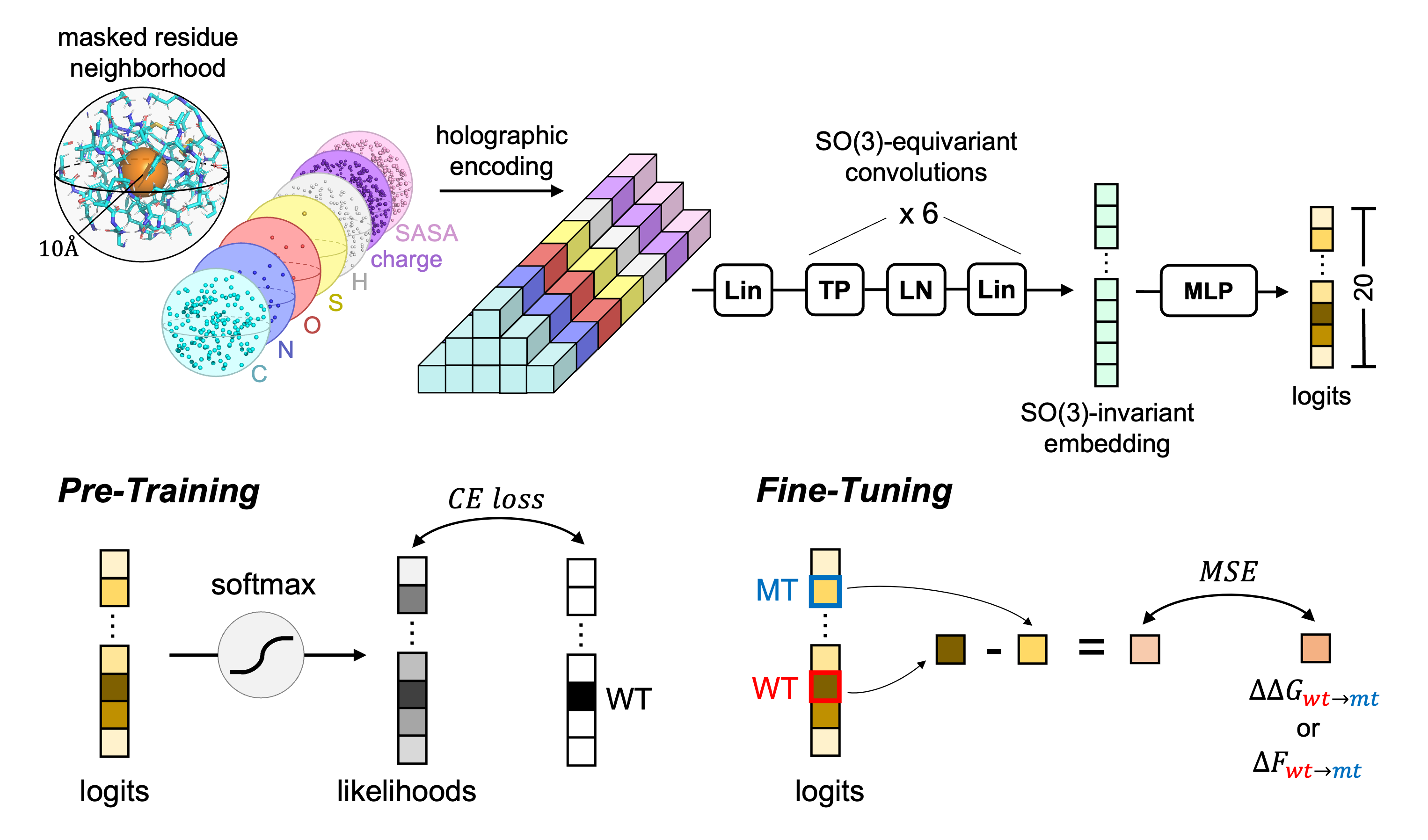
Code for the paper HERMES: Holographic Equivariant neuRal network model for Mutational Effect and Stability prediction.
You can run predictions easily on the Google Colab Notebook.
NOTE: Currently, there is a conflict between openmm and pytorch, whereby it's challenging to install both with CUDA support. We are currently struggling to replicate the installation on our local HPC cluster - which we once managed to do (sigh...). Install pytorch with CPU support only, however, gives no issues. Try installing pytorch with CUDA first, and if that doesn't work, install it with CPU only. We are working on a foolproof solution - if you have any leads please let us know by posting an Issue.
Step 1: Create environment and install pytorch.
conda create -n protholo python=3.9
conda activate protholoInstall pytorch==1.13.1 with or without CUDA depending on whether you have a GPU available, following https://pytorch.org/get-started/previous-versions/. For example:
conda install pytorch==1.13.1 torchvision==0.14.1 torchaudio==0.13.1 pytorch-cuda=11.7 -c pytorch -c nvidia # with cuda for gpu support
conda install pytorch==1.13.1 torchvision==0.14.1 torchaudio==0.13.1 cpuonly -c pytorch # cpu only, use this if having issues with step 2Step 2: Install zernikegrams package, which we use for protein preprocessing.
conda install zernikegrams -c statphysbio -c conda-forgeTD;DR: if you are experiencing issues with the installation and you have pytorch with CUDA installed, install pytorch with CPU only.
Installing zernikegrams will also install other necessary packages such as openmm. As outlined above, in our environment we are experiencing a conflict between openmm and pytorch that we are currently working on resolving, and cannot guarantee GPU support at this time (though it works without issues on colab). \
(Optional) Step 3: Install pyrosetta. This is required for the use of models trained on structures processed using pyrosetta. A license is available at no cost to academics and can be obtained here. We are aware of the limitations posed by pyrosetta's license and are working on releasing a version that uss biopython instead and other open source code soon.
To download pyrosetta, after obtaining a license from the link above, follow instructions here. We recommend downloading the .whl file and installing with pip. Our models were trained and evaluated using version PyRosetta4.Release.python39.linux.release-335. We do not foresee issues with using alternative versions, but cannot guarantee compatibility at this time.
Step 4: Install protein_holography_web as a package. This will install some of the other necessary packages as well.
pip install .Feel free to use pip install -e . instead if you plan on making changes to the code and want to test them without reinstalling the package.
Installation tips:
- The error
{ENVIRONMENT_PATH}/bin/../lib/libstdc++.so.6: version 'GLIBCXX_3.4.30' not found, as required by OpenMM, can be fixed viaconda install -c conda-forge libstdcxx-ng. See https://stackoverflow.com/questions/48453497/anaconda-libstdc-so-6-version-glibcxx-3-4-20-not-found - Somehow can't install pdbfixer AFTER conda
We are in the process of re-naming models in the repository. For now, please use the old names.\ [new name] <--> [old name]
We provide the following pre-trained models:
HERMES_Bp_000<-->HCNN_biopython_proteinnet_extra_mols_0p00: trained on ~10k CASP12 ProteinNet chains, using the preprocessing pipeline from the RaSP paper based on Biopython, except we keep extra ligands and ions (not water).HERMES_Bp_050<-->HCNN_biopython_proteinnet_extra_mols_0p50: same as above, but trained on data with added gaussian noise with standard deviation of 0.5 Angstrom.HERMES_Py_000<-->HCNN_pyrosetta_proteinnet_extra_mols_0p00: trained on ~10k CASP12 ProteinNet chains, using pyrosetta to preprocess protein structures as described in the HCNN paper. This procedure includes ligands and ions, excludes water, and - constrary to RaSP - does not substitute non-canonical amino-acids.HERMES_Py_050<-->HCNN_pyrosetta_proteinnet_extra_mols_0p50: same as above, but trained on data with added gaussian noise with standard deviation of 0.5 Angstrom.
We also provide the same models fine-tuned on stability ddG values computed with Rosetta:
HERMES_Bp_000_FT_Ros_ddG<-->HCNN_biopython_proteinnet_extra_mols_0p00_finetuned_with_rosetta_ddg_allHERMES_Bp_050_FT_Ros_ddG<-->HCNN_biopython_proteinnet_extra_mols_0p50_finetuned_with_rosetta_ddg_with_0p50_allHERMES_Py_000_FT_Ros_ddG<-->HCNN_pyrosetta_proteinnet_extra_mols_0p00_finetuned_with_rosetta_ddg_allHERMES_Py_050_FT_Ros_ddG<-->HCNN_pyrosetta_proteinnet_extra_mols_0p50_finetuned_with_rosetta_ddg_with_0p50_all
Please see the paper for a comprehensive benchmarking evaluation. The tl;dr is: fine-tuned models work better, and all four perform similarly. Pre-trained-only models work better with 0.50 Angstrom noise, and with pyrosetta preprocessing.
Note that, to use the pyrosetta models, a local installation of pyrosetta is necessary.
The script run_hcnn_on_pdbfiles.py can be given as input a set of PDB files - with optionally pdb-specific chains - and it will output a csv file where every row is a uniquely-identified site, and columns are the site's mutation probabilities. If embeddings are requested, they will be outputted in a separate file called {CSV_FILENAME}-embeddings.npy.
usage: run_hcnn_on_pdbfiles.py [-h] -m MODEL_VERSION [-hf HDF5_FILE] [-pd FOLDER_WITH_PDBS] [-pn FILE_WITH_PDBIDS_AND_CHAINS] [-pp PARALLELISM] -o OUTPUT_FILEPATH [-r {logprobas,probas,embeddings,logits} [{logprobas,probas,embeddings,logits} ...]] [-an {0,1}] [-el {0,1}] [-bs BATCH_SIZE] [-v {0,1}] [-lb {0,1}]
optional arguments:
-h, --help show this help message and exit
-m MODEL_VERSION, --model_version MODEL_VERSION
Name of HCNN model you want to use.
-hf HDF5_FILE, --hdf5_file HDF5_FILE
Path to an .hdf5 file containing zernikegrams and res_ids to run inference on. Cannot be specified together with --folder_with_pdbs.
-pd FOLDER_WITH_PDBS, --folder_with_pdbs FOLDER_WITH_PDBS
Directory containing PDB files to run inference on. Inference is run on all sites in the structure. Cannot be specified together with --hdf5_file.
-pn FILE_WITH_PDBIDS_AND_CHAINS, --file_with_pdbids_and_chains FILE_WITH_PDBIDS_AND_CHAINS
[Optional] Path to a .txt file containing pdbids and chains to run inference on. If not specified, and --folder_with_pdbs is specified, inference will be run on all sites in the structure. If specified, each line should be in the format "pdbid chain"; if chain is not specified for a given line, inference will be run on all chains in that
structure.
-pp PARALLELISM, --parallelism PARALLELISM
If zero (default), pdb files are processed one by one. If one, pdb files are processed in parallel with specified parallelism (and number of cores available), by first generating zernikegrams in a temporary hdf5 file.
-o OUTPUT_FILEPATH, --output_filepath OUTPUT_FILEPATH
Must be a ".csv file". Embeddings will be saved separately, in a parallel array, with the same filename but with the extension "-embeddings.npy".
-r {logprobas,probas,embeddings,logits} [{logprobas,probas,embeddings,logits} ...], --request {logprobas,probas,embeddings,logits} [{logprobas,probas,embeddings,logits} ...]
Which data to return. Can be a combination of "logprobas", "probas", "embeddings", and "logits".
-an {0,1}, --add_same_noise_level_as_training {0,1}
1 for True, 0 for False. If True, will add the same noise level as was used during training. This is useful for debugging purposes. Default is False.
-el {0,1}, --ensemble_at_logits_level {0,1}
1 for True, 0 for False. When computing probabilities and log-probabilities, ensembles the logits before computing the softmax, as opposed to ansembling the individual models' probabilities. There should not be a big difference, unless the ensembled models are trained very differently.
-bs BATCH_SIZE, --batch_size BATCH_SIZE
Batch size for the model (number of sites). Higher batch sizes are faster, but may not fit in memory. Default is 512.
-v {0,1}, --verbose {0,1}
0 for no, 1 for yes. Currently, "yes" will print out accuracy of the model on the data.
-lb {0,1}, --loading_bar {0,1}
0 for no, 1 for yes.Some common use cases:
Get all probabilities and embeddings for all sites in the PDB files found in pdbs.
python run_hcnn_on_pdbfiles.py -pd pdbs --m HCNN_biopython_proteinnet_0p00 -o all_sites.csv -r probas embeddingsThe above command will output two csv files: one called all_sites.csv with mutation probabilities, the other called all_sites-embeddings.npy with embeddings, for all sites in the PDB files found in the pdbs directory.
Request to process specific pdbs and chains.
The requested pdbs and chains should be listed in a text file, with one pdb and, optionally, a single chain per line. If the chain is not specified, then all chains are considered. To consider multiple chains, list the pdb multiple times with each different chain. Note that we DO NOT delete the atoms of the non-requested chains.
For example, assume the file my_pdbs_and_chains.txt contains the following:
1ao7
1qrn A
1qrn B
Then, to process only these pdbs and chains, run:
python run_hcnn_on_pdbfiles.py -pd pdbs -pn my_pdbs_and_chains.txt --m HCNN_biopython_proteinnet_0p00 -o specific_chains.csv -r probas embeddingsIf a requested pdb file is not found in the directory, the script will automatically attempt to download it from the RCSB website.
New from 08/05/24 - Parallel processing with multiprocessing.
We now support processing proteins in parallel over multiple cores, leveraging the multiprocessing library. To use this feature, first make sure your program has access to the desired number of cores, then provide the -pp argument with the number of cores you want to use. Crucially, this will parallelize only the processing and parsiong of proteins, not the forward pass to the model. Furthermore, note that multiprocessing has some significant overhead, so the time improvement is less than linear with the number of cores and, in some cases, using multiprocessing might even be slower than not parallelizing at all. If you do not want to use multiprocessing, use the default value -pp 0, as specifying -pp 1 will still call multiprocessing and be slower. \
Below are the times to run HERMES_Bp_000 on 15 PDBs using a single A40 GPU, and access to 5 cores with 64GB total of memory, and with varying degrees of parallelization:
-pp |
Time (s) |
|---|---|
| 0 | 514 |
| 1 | 628 |
| 2 | 367 |
| 3 | 332 |
You can run this test yourself by running the function test_run_hcnn_on_pdbfiles__with_parallelism_and_without() from 'tests/test_run_hcnn_on_pdbfiles.py'.
New from 08/05/24 - Parallel processing with SLURM.
A more efficient option for parallel processing, which however requires more code to set up, is to call the script run_hcnn_on_pdbfiles.py in parallel on subsets of the PDB files and then merge the results. This is most convenient when using a job scheduler like SLURM. We provide a script that automatically runs all HERMES models on all PDB files in a directory, by submitting a single-core job per PDB-model combination. It is the responsibility of the user to then merge the results if they so desire. The script it easily modifiable and we invite the experienced users to modify it to their needs. The script is called run_hcnn_on_pdbfiles_parallel_with_slurm.py:
usage: run_hcnn_on_pdbfiles_in_parallel_with_slurm.py [-h] [-pd FOLDER_WITH_PDBS] [-df DUMPFILES_FOLDER] [-of OUTPUT_FOLDER] [-hf HERMES_FOLDER] [-A ACCOUNT] [-P PARTITION] [-G {0,1}] [-C NUM_CORES] [-W WALLTIME] [-M MEMORY] [-E {0,1}] [-EA EMAIL_ADDRESS]
optional arguments:
-h, --help show this help message and exit
-pd FOLDER_WITH_PDBS, --folder_with_pdbs FOLDER_WITH_PDBS
Directory containing PDB files to run inference on. Inference is run on all sites in the structure.
-df DUMPFILES_FOLDER, --dumpfiles_folder DUMPFILES_FOLDER
Root to store dumpfiles.
-of OUTPUT_FOLDER, --output_folder OUTPUT_FOLDER
Root to store outputs.
-hf HERMES_FOLDER, --hermes_folder HERMES_FOLDER
Path to the Hermes folder, containing the run_hcnn_on_pdbfiles.py script.
-A ACCOUNT, --account ACCOUNT
-P PARTITION, --partition PARTITION
-G {0,1}, --use_gpu {0,1}
-C NUM_CORES, --num_cores NUM_CORES
-W WALLTIME, --walltime WALLTIME
-M MEMORY, --memory MEMORY
-E {0,1}, --send_emails {0,1}
-EA EMAIL_ADDRESS, --email_address EMAIL_ADDRESSSometimes, it is useful to score specific mutations. The script zero_shot_mutation_effect_prediction_with_hcnn.py can be used for this purpose. It takes as input a csv file with columns corresponding to: the mutation, the chain, and the pdb file of the wildtype. The script will output a csv file with the mutation probabilities and embeddings for the requested mutations.
If desired, the script supports the use of the mutant structure to predict the mutation effect. This can be done by providing the mutant pdb file in the csv file in the appropriate column.
The columns are not expected to have specific names, but the names must ben provided as input to the script.
Run python zero_shot_mutation_effect_prediction_with_hcnn.py -h for more information on the script, and see experiments/Protein_G/ for a simple example.
Fine-tuning can be easily done in three steps.
-
Prepare the data. Prepare the targets in three .csv files, which must have
{train},{valid}, and{test}in the name. Each .csv file must have the following columns:[pdbid, chainid, variant, score]. Also, place all the pdbfiles for training, validation and testing in a single directory. -
Generate inputs (aka zernikegrams or holograms). For faster training, we pre-generate the inputs and store them in a .npz file. Run
make_zernikegrams_for_finetuning.pyto generate the inputs, providing as arguments, the model you want to make inputs for, the directory of pdbfiles, whether to add noise to structures, and the output directory. -
Fine-tune the model. Run
finetune_hcnn.pyto fine-tune the model. You need to provide a config file with the necessary information, see/training_data/finetuning/rosetta_ddg/configs/HCNN_biopython_proteinnet_extra_mols_0p00__all.yamlfor a thorough example.
If you use this code, please cite the following paper:
@article {visani_hermes_2024,
author = {Visani, Gian Marco and Pun, Michael N. and Galvin, William and Daniel, Eric and Borisiak, Kevin and Wagura, Utheri and Nourmohammad, Armita},
title = {HERMES: Holographic Equivariant neuRal network model for Mutational Effect and Stability prediction},
elocation-id = {2024.07.09.602403},
year = {2024},
doi = {10.1101/2024.07.09.602403},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2024/07/13/2024.07.09.602403},
eprint = {https://www.biorxiv.org/content/early/2024/07/13/2024.07.09.602403.full.pdf},
journal = {bioRxiv}
}