A database of paths that represent the mechanism of action from a drug to a disease in an indication. A preliminary relase of DrugMechDB was published here:
Database of mechanism of action paths for selected drug-disease indications
Mayers, Michael; Steinecke, Dylan; Su, Andrew I.
A web interface to DrugMechDB can be found at https://sulab.github.io/DrugMechDB/.
Most drug mechanisms are described simply as a protein target, or sometimes a pathway, that then treats the disease. However, there are usually more steps across a biological knowledge graph required to traverse from the target through its mechanism to the reduction of a disease. This database attempts to catalog a subset of known drug-disease indications as a path through a network of biomedical entities.
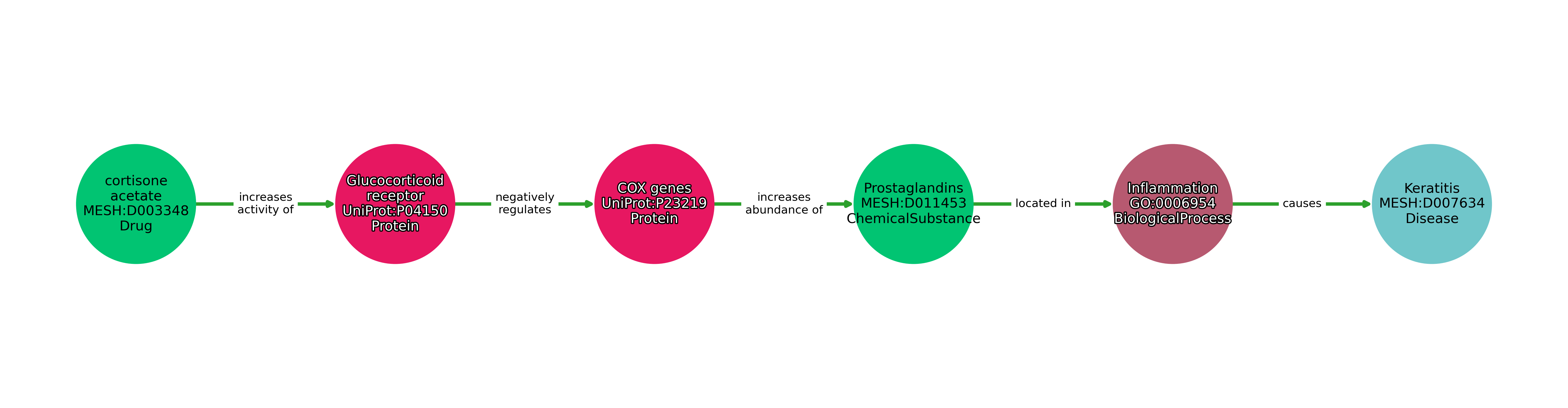
Below is a visualized example of one entry in DrugMechDB - A path from Cortisone acetate to Keratitis.
Indications were selected at random from the set contained within DrugCentral. Half were taken from the full set, representing a set of common drug disease pairs, and the other half were selected from a pool of less-common diseases to vary the targets and diseases treated.
Paths were derived from free-text descriptions found on DrugBank, Wikipeida and within literature. Concepts within the text were then normalized to a concept type (Drug, Protein, Pathway, etc) and relationships between the concepts were determined from the source. Finally concepts were mapped to an identifier depending on the concept type according to the following table:
If you would like to contribute your own curated mechanistic paths, please do so by making pull requests
with edits to the file submission.yaml.
See the Curation Guide for more information about contributions and Submission Guide for detailed submission instructions.
Each path begins with a - directed: true statment. Identifiers for concepts and concept type
should conform to the table above.
Paths contain the following structure:
- directed: true
graph:
disease: *name of the disease in the indication*
disease_mesh: *MESH Identifier for the disease (if known)*
drug: *name of the drug in the indication*
drug_mesh: *MESH Identifier for the drug (if known)*
drugbank: *DrugBank Identifier for the drug (if known)*
links: (the edges of the path)
- key: *Semantics of the relationship (ALL CAPS)*
source: *Identifier for source node in edge*
target: *Identifier for target node in edge*
nodes: (the nodes in the path)
- id: *Identifier for the node*
label: *Concept type for the node*
name: *Name of the node*
multigraph: true (required statment for importing paths into networkx).
We originally had this configured so that the "Update Website" action would build the Jekyll files and push to the gh-pages branch, which would then be rendered by Github Pages. However, the web content is now too large and the build process times out. Therefore, we have to build locally. The protocol is as follows:
-
Execute the "Merge Pages" action which will merge the
mainbranch togh-pages -
Create the
gh-pagesbranch locallygit clone --branch gh-pages --depth 1 git@github.com:SuLab/DrugMechDB.git DrugMechDB.gh-pages cd DrugMechDB.gh-pagesor if the repo already exists locally,
cd DrugMechDB.gh-pages git pull -
Create the Jekyll files from
indication_paths.yamlpython parse.py -
Use jekyll to render HTML output:
This command can take up to ~30 mins to complete, and output HTML files will be in
_sitebundle exec jekyll build -
Create the
gh-pages-htmlbranch locally:cd .. git clone --branch gh-pages-html git@github.com:SuLab/DrugMechDB.git DrugMechDB.gh-pages-html cd DrugMechDB.gh-pages-htmlor if the repo already exists locally,
cd DrugMechDB.gh-pages-html git pull -
Copy the new rendered files:
rm -rf * cp -r ../DrugMechDB.gh-pages/_site/* . -
Commit and push:
git add . git commit -m 'Update website' git push