This repo contains PyTorch implementation of the paper "Learning Neural Set Functions Under the Optimal Subset Oracle"
by Zijing Ou, Tingyang Xu, Qinliang Su, Yingzhen Li, Peilin Zhao, and Yatao Bian.
We propose a way to learn set functions when the optimal subsets are given from an optimal subset oracle. This setting is different to other works that learn set functions from the function value oracle that provide utility values for each specific subset. Thus this setting is arguably more practically important but is surprisingly overlooked by previous works. To learn set functions under the optimal subset oracle, we propose to cast the problem into maximum likelihood estimation by replacing the utility function with an energy-based model such that it is proportional to the utility value, and satisfies some desiderata for set functions (e.g., permutation invariance, etc). Then mean-field variational inference and its amortized variants are proposed to learn EBMs on the sets. We evaluate our approach in a wide range of applications, including product recommendation, set anomaly detection, and compound selection in AI-aided drug discovery. The empirical results show our approach is promising.
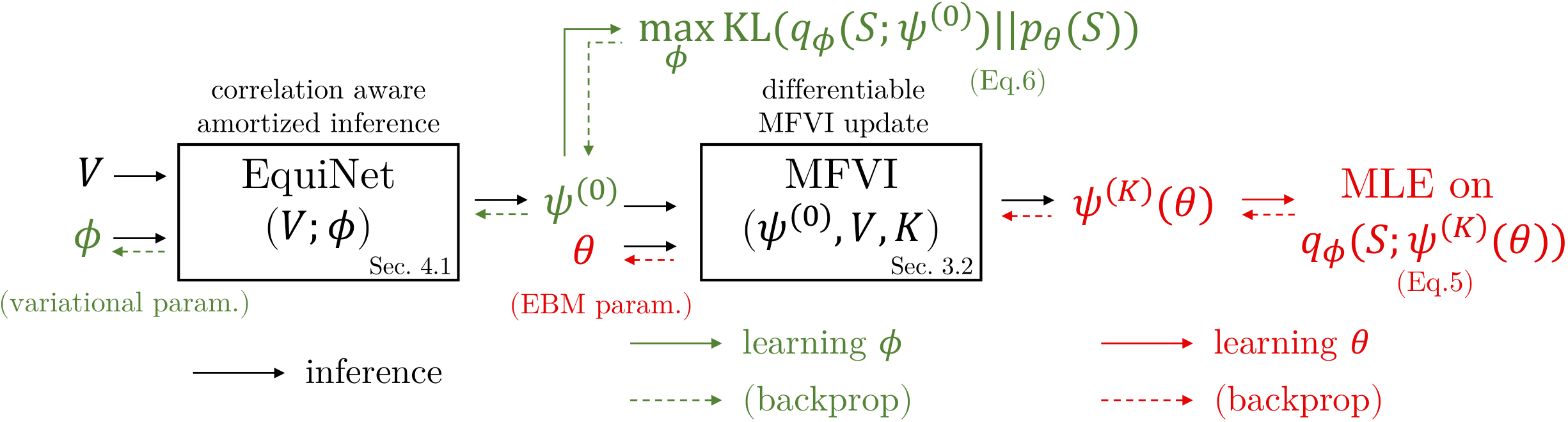
Overview of the training and inference processes of EquiVSet.
Please ensure that:
- Python >= 3.6
- PyTorch >= 1.8.0
- dgl >= 0.7.0
The following pakages are needed if you want to run the compound selection experiments:
- rdkit: We recommend installing it with
conda install -c rdkit rdkit==2018.09.3. For other installation recipes, see the official documentation. - dgllife: We recommend installing it with
pip install dgllife. More information is available in the official documentation. - tdc: We recommend installing it with
pip install PyTDC. See the official documentation for more information.
We provide step-by-step installation commands as follows:
conda create -n EquiVSet python=3.7
source activate EquiVSet
pip install torch==1.9.0+cu111 torchvision==0.10.0+cu111 torchaudio==0.9.0 -f https://download.pytorch.org/whl/torch_stable.html
pip install dgl-cu110 dglgo -f https://data.dgl.ai/wheels/repo.html
# The following commands are used for compound selection:
conda install -c rdkit rdkit==2018.09.3
pip install dgllife
pip install PyTDC
For the experiments, we use the following datasets:
- Amazon baby registry dataset for the
product recommendationexperiments. The dataset is available here. - CelebA dataset fot the
set anomaly detectionexperiments. The images are available here and the attribute labels are available here. - PDBBind and BindingDB for the
compubd selectionexperiments. The PDBBind dataset is available here and the BindingDB dataset is available here.
For all experiments, the dataset is automatically downloaded and preprocessed when you run the corresponding code. You could also download the dataset manually using the link provided.
This repository implements the synthetic experiments (section 6), product recommendation (section 6), set anomaly detection (section 6), and compound selection (section 6).
To run on the Two-Moons and Gaussian-Mixture dataset
python main.py equivset --train --cuda --data_name <dataset_name>
dataset_name is chosen in ['moons', 'gaussian'].
We also provide the Jupyter notebook to run the synthetic experiments.
To run on the Amazon baby registry dataset
python main.py equivset --train --cuda --data_name amazon --amazon_cat <category_name>
category_name is chosen in ['toys', 'furniture', 'gear', 'carseats', 'bath', 'health', 'diaper', 'bedding', 'safety', 'feeding', 'apparel', 'media'].
To run on the CelebA dataset
python main.py equivset --train --cuda --data_name celeba
To run on the PDBBind and BindingDB dataset
python main.py equivset --train --cuda --data_name <dataset_name>
dataset_name is chosen in ['pdbbind', 'bindingdb'].
😄If you find this repo is useful, please consider to cite our paper:
@article{ou2022learning,
title={Learning Set Functions Under the Optimal Subset Oracle via Equivariant Variational Inference},
author={Ou, Zijing and Xu, Tingyang and Su, Qinliang and Li, Yingzhen and Zhao, Peilin and Bian, Yatao},
journal={arXiv preprint arXiv:2203.01693},
year={2022}
}