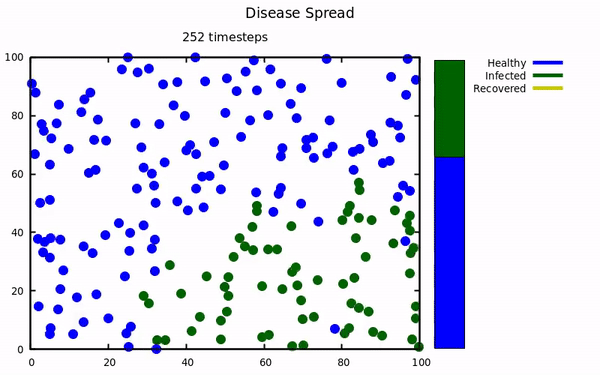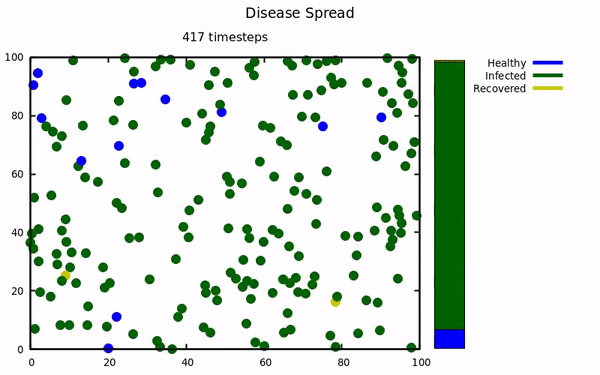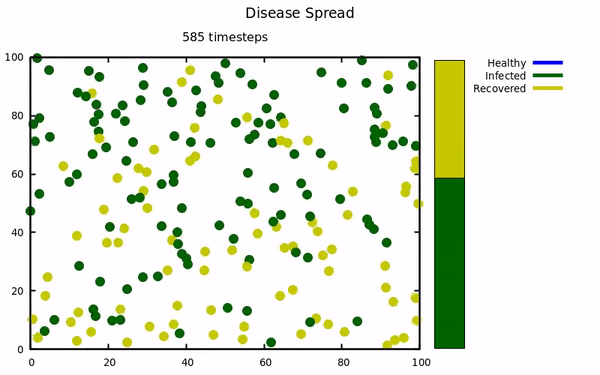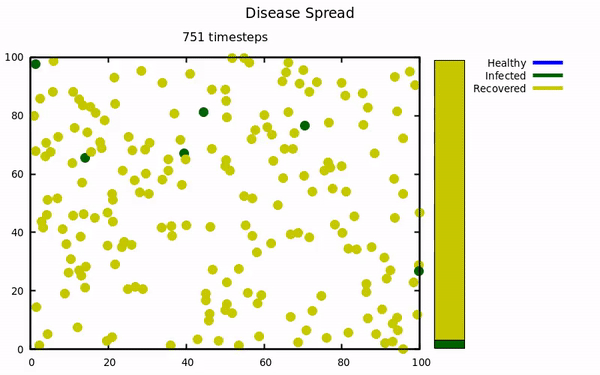
An simulation using molecular dynamics to model infectious spread among small populations.
At the start of the pandemic, I was taking a computational physics course. Our instructor emailed the class a link to a COVID-19 simulation that implemented molecular dynamics to model spread. This project was undertaken to try and replicate their results with my own simulation. The Physics isn't exactly comprehensive (energy is not conserved, collisions aren't elastic, no molecular attractive/repulsive forces, etc...) because those characteristics were not present in the original simulation.
- C++
- Makefile
- Gnuplot
- FFMPEG
I've only run this using Linux, so I'm kind of unfamiliar with setup on Windows, but I recommend setting up WSL and following the Linux installation below if you really wanna check it out.
The versions of ffmpeg and gnuplot given below are just the versions I used; I'm sure any other major versions would also work (e.g. make sure gnuplot is at least major version 5).
# Clone the repo
$ git clone https://github.com/Svaught598/Infection-Sim.git
$ cd Infection-Sim
# Install make and g++ (if you don't already have them)
$ sudo apt-get install make
$ sudo apt-get install g++
# Install ffmpeg and gnuplot
$ sudo apt-get install ffmpeg=7:4.2.4-1ubuntu0.1
$ sudo apt-get install gnuplot=5.2.8+dfsg1-2
# Compile and run!
$ make runMIT License