Szewczyk TM, T Lee, MJ Ducey, ME Aiello-Lammens, H Bibaud, JM Allen. 2019. Local management in a regional context: Simulations with process-based species distribution models. Ecological Modelling 413(1): 108827
Code repository accompanying Szewczyk et al. 2019
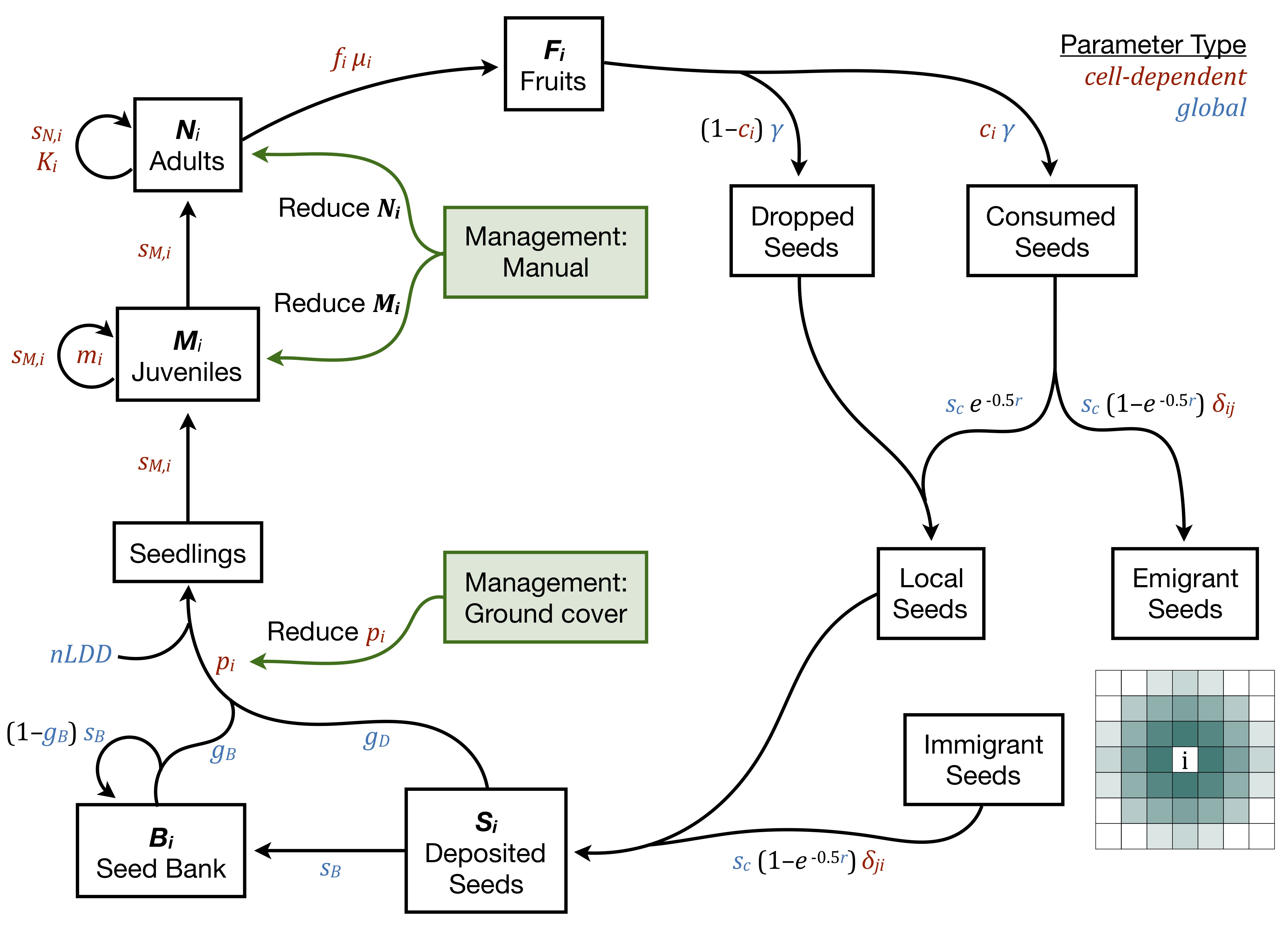
This R package runs a population-level demographic cellular automata model. The life history structure and default parameterization are based on glossy buckthorn (Frangula alnus) in New England. The model is spatially explicit and temporally dynamic, simulating the growth and spread of a species on a gridded landscape. Population and life history dynamics occur within each occupied grid cell, driven by vital rates and demographic parameters which can be global or dependent on the cell environment. Cells are connected through mechanistic short distance dispersal (SDD) and rare long distance dispersal (LDD). Management actions can target specific life stages, such as reducing seedling establishment (e.g., through planted ground cover) or reducing the number of adults (e.g., through cutting and/or spraying). In each year, the simulation occurs according to the following series of steps:
with the following parameters and containers for each cell i, each time t, and juvenile ages k = 1:(m-1):
| Symbol | Quantity |
|---|---|
| Nit | Number of adults |
| Mitk | Number of juveniles |
| Fit | Number of fruits |
| Sit | Number of seeds deposited |
| Bit | Number of seeds in seed bank |
| fi | Mean flowering probability |
| μi | Mean number of fruits from flowering adult |
| γ | Mean number of seeds per fruit |
| mi | Age at adulthood |
| ci | Pr(fruit is consumed by a bird) |
| r | Rat parameter for SDD exponential kernel (units: cells) |
| sddmax | Maximum SDD distance (units: cells) |
| Ji | Number of SDD neighbors (calculated from sddmax) |
| δji | Pr(emigrant from j is deposited in i) |
| nldd | Number of LDD events |
| sc | Pr(seed survival | consumed) |
| sB | Pr(seed survival in the seed bank) |
| sM,i | Pr(juvenile survival) |
| sN,i | Pr(adult survival) |
| Ki | Carrying capacity for adults |
| gD | Pr(germinating directly: same year as produced) |
| gB | Pr(germinating from the seed bank) |
| pi | Pr(establishing once germinated) |
Note that in the code, the ages 1:(m-1) are included as layers in the object N rather than as a separate object M.
Parameters that vary with among cells can be estimated through regressions with environmental covariates (Merow et al 2017) as appropriate for the species and system. Density dependence can be implemented either as a hard cap on the abundance within each cell (Merow et al 2011) or through parameters such as the establishment probability, survival probabilities, or fruiting probabilities (Ellner & Rees 2006).
The model is run using the function run_sim(), though some set up is necessary for initializing the landscape. The script vignettes/management_controls.R assigns parameter values, establishes management strategies, and builds the landscape before running a set number of stochastic simulations. The script hpc/hpc_wrap.R runs a global sensitivity analysis, where many parameters are varied simultaneously.