The organ.ipynb notebook is a Python script designed for processing and analyzing medical imaging data. It uses a variety of packages to load, transform, and visualize this data, with a particular focus on DICOM files, which are a standard format for medical imaging data. Probably the most important element of this project is the MONAI framework which provides a robust set of tools in AI development for healthcare researchers, with an immediate focus on medical imaging.
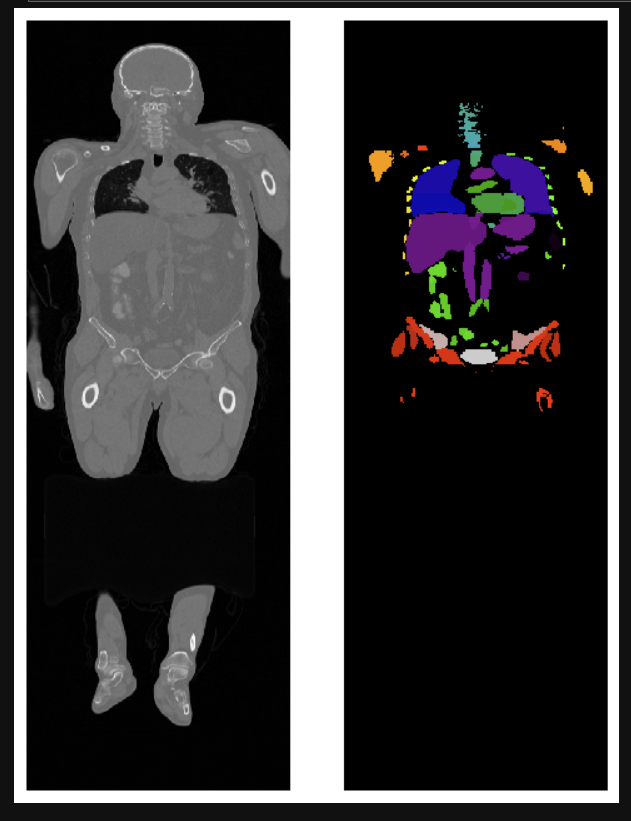
Image segmentation has become a key process for the delineation of certain anatomical structures and other regions to assist and aid physicians in surgery, biopsies, and other clinical tests.
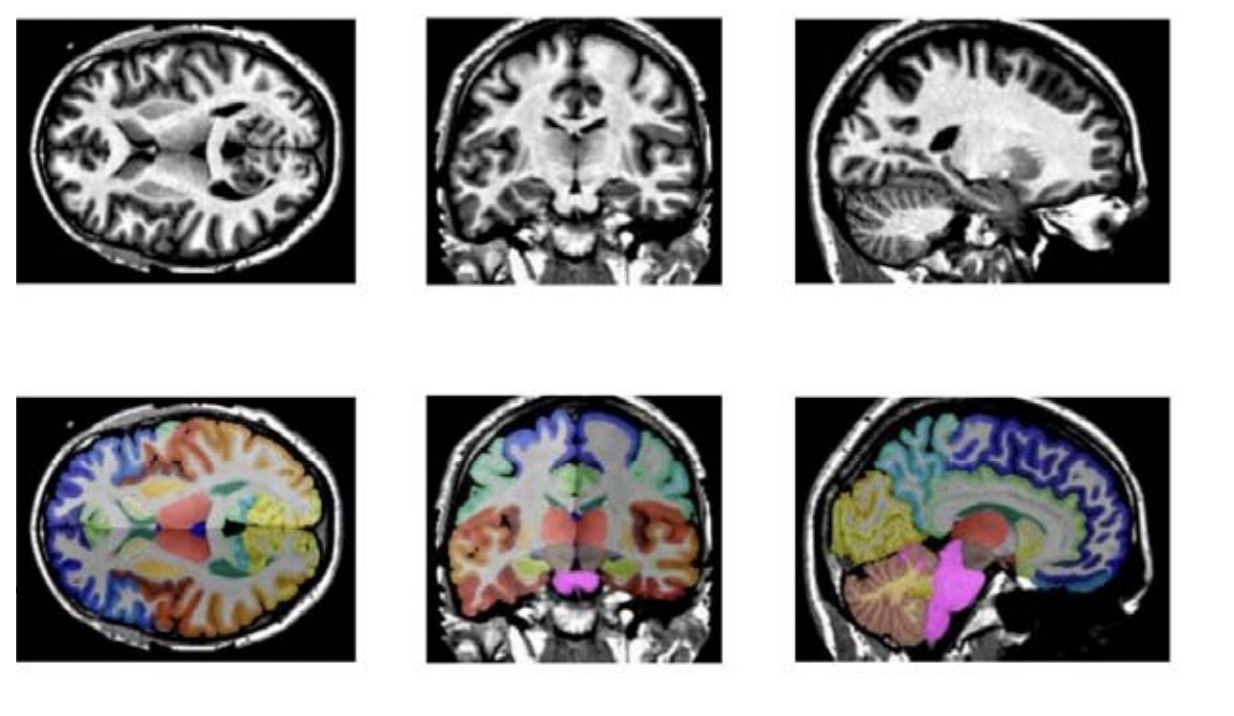
The image below is from a detailed whole brain segmentation. It is an essential quantitative technique in medical image analysis, which provides a non-invasive way of measuring brain regions from a clinical acquired structural magnetic resonance imaging (it's not just CT imaging).
Credit to: Xin Yu (xin.yu@vanderbilt.edu) and Yinchi Zhou (yinchi.zhou@vanderbilt.edu) | Yucheng Tang (yuchengt@nvidia.com)
This notebook requires the following Python packages:
- os
- numpy
- torch
- pydicom: A Python package specifically for parsing and manipulating DICOM files.
- matplotlib:
- tcia_utils: A custom module for interacting with The Cancer Imaging Archive (TCIA).
- monai: A PyTorch-based framework providing tools and components to build and train neural networks for medical imaging tasks.
- rt_utils: A utility for handling RT Dose, RT Structure Set, and RT Plan DICOM files.
- scipy: A Python library used for scientific and technical computing.
This notebook requires Python 3.9.0. Some of the dependencies do not yet support Python 3.10, so it's important to use Python 3.9.0 to avoid compatibility issues. It took me one whole day to find the proper Python version so you don't need to.
To install the required packages, refer to my requirements.txt file.
To use this notebook, open it in Jupyter Notebook and run the cells in order. The notebook includes comments and markdown cells that explain what each part of the code does. Once you have decided which CT image you'd want to use from the TCIA or any other public library you can go ahead and follow the steps in the notebook. I've downloaded a whole body CT scan which is in my Data folder.