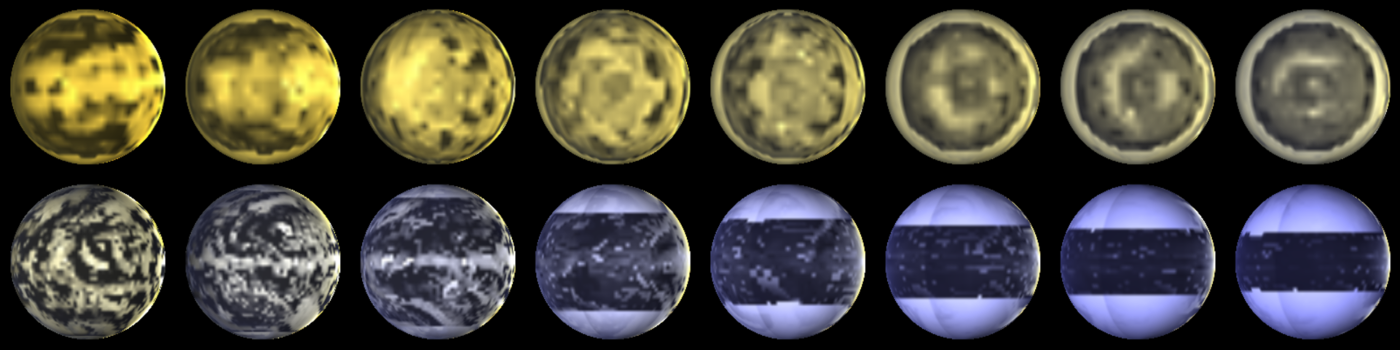
 A range of planets modeled by ExoPlaSim, and postprocessed with SBDART. The top row consists of tidally-locked aquaplanets at T21 orbiting stars ranging from 2500 K to 4000 K, with orbital periods increasing with stellar mass. The bottom row consists of aquaplanets with 24-hour rotation at T42, orbiting stars ranging from 4000 K to 8000 K.
A range of planets modeled by ExoPlaSim, and postprocessed with SBDART. The top row consists of tidally-locked aquaplanets at T21 orbiting stars ranging from 2500 K to 4000 K, with orbital periods increasing with stellar mass. The bottom row consists of aquaplanets with 24-hour rotation at T42, orbiting stars ranging from 4000 K to 8000 K.
The PlaSim 3D general climate model, extended for terrestrial planets. This model contains the PlaSim GCM, as well as all necessary modifications to run tidally-locked planets, planets with substantially different surface pressures than Earth, planets orbiting stars with different effective temperatures, super-Earths, and more. This model also includes the ability to compute carbon-silicate weathering, dynamic orography through the glacier module (though only accumulation and ablation/evaporation/melting are included; glacial flow and spreading are not), and storm climatology. Future features will include support for multiple celestial light sources (e.g. for a habitable moon orbiting a Jovian planet, or circumbinary planets), coupling with N-body integrators such as REBOUND, and CO2 condensation.
This model ships with a Python API, described below. Full documentation of the API is available at http://exoplasim.readthedocs.io.
Documentation of the original PlaSim model is found in exoplasim/plasim/doc.
Created by Adiv Paradise
Copyright 2020, Distributed under the General Public License
This API was written with Python 3 in mind, but should work with Python 2 and outdated versions of NumPy.
- numpy
- scipy
- matplotlib (only needed for additional utilities)
- GNU C (gcc/g++) and Fortran (gfortran) compilers (for Python utilities)
- (optionally) Other compilers whose use you prefer for the model itself
- (optionally) MPI libraries for those compilers
- netCDF4 (optional)
- h5py (optional)
Compatibility
- Linux (tested on Ubuntu 18.04, CentOS 6.10): Yes
- Google Colaboratory: Yes (note that OpenMPI support on Colaboratory is limited due to automatic root privileges; look up how to run OpenMPI executables with root permissions and note that this is not recommended)
- Windows 10: Yes, via Windows Subsystem for Linux
- Mac OS X: Yes, requires Xcode and developer tools, and OpenMPI support requires that Fortran-compatible libraries be built. Tested on Mac OS X Catalina and Big Sur (with MacPorts, GCC10, OpenMPI, and Anaconda3), Apple M1 compatibility has not been tested.
- ExoPlaSim no longer depends on X11 libraries for installation and compilation!
- Revamped postprocessor no longer depends on NetCDF-C libraries, and supports additional output formats (including netCDF, HDF5, NumPy archives, and archives of CSV files).
- GCC and gfortran support through GCC 10.
- Improved cross-platform compatibility
- Numerous bugfixes
pip install exoplasim
OR:
python setup.py install
The first time you import the module and try to create a model after either installing or updating, ExoPlaSim will run a configuration script.
Multiple output formats are supported by the built-in pyburn
postprocessor. If you wish to use HDF5 or NetCDF output formats, you
will need the netCDF4-python and h5py libraries, respectively. You
can ensure these are included at install-time by specifying them:
pip install exoplasim[netCDF4]
OR:
pip install exoplasim[HDF5]
OR:
pip install exoplasim[netCDF4,HDF5]
You may also configure and compile the model manually if you wish to not
use the Python API, by entering the exoplasim/ directory and running
first configure.sh, then compile.sh (compilation flags are shown by
running ./compile.sh -h).
Original PlaSim documentation is available in the exoplasim/docs/ folder.
To use the ExoPlaSim Python API, you must import the module, create a Model or one of its subclasses, call its configure method and/or modify method, and then run it.
An IPython notebook is included with ExoPlaSim; which demonstrates basic usage. It can be found in the ExoPlaSim installation directory, or downloaded directly here.
Basic example::
import exoplasim as exo
mymodel = exo.Model(workdir="mymodel_testrun",modelname="mymodel",resolution="T21",layers=10,ncpus=8)
mymodel.configure()
mymodel.exportcfg()
mymodel.run(years=100,crashifbroken=True)
mymodel.finalize("mymodel_output")
In this example, we initialize a model that will run in the directory
"mymodel_testrun", and has the name "mymodel", which will be used to
label output and error logs. The model has T21 resolution, or 32x64, 10
layers, and will run on 8 CPUs. By default, the compiler will use 8-byte
precision. 4-byte may run slightly faster, but possibly at the cost of
reduced stability. If there are machine-specific optimization flags you
would like to use when compiling, you may specify them as a string to
the optimization argument, e.g. optimization='mavx'. ExoPlaSim will
check to see if an appropriate executable has already been created, and
if not (or if flags indicating special compiler behavior such as
debug=True or an optimization flag are set) it will compile one. We then
configure the model with all the default parameter choices, which means
we will get a model of Earth. We then export the model configurations to
a .cfg file (named automatically after the model), which will allow
the model configuration to be recreated exactly by other users. We run
the model for 100 years, with error-handling enabled. Finally, we tell
the model to clean up after itself. It will take the most recent output
files and rename them after the model name we chose, and delete all the
intermediate output and configuration files.