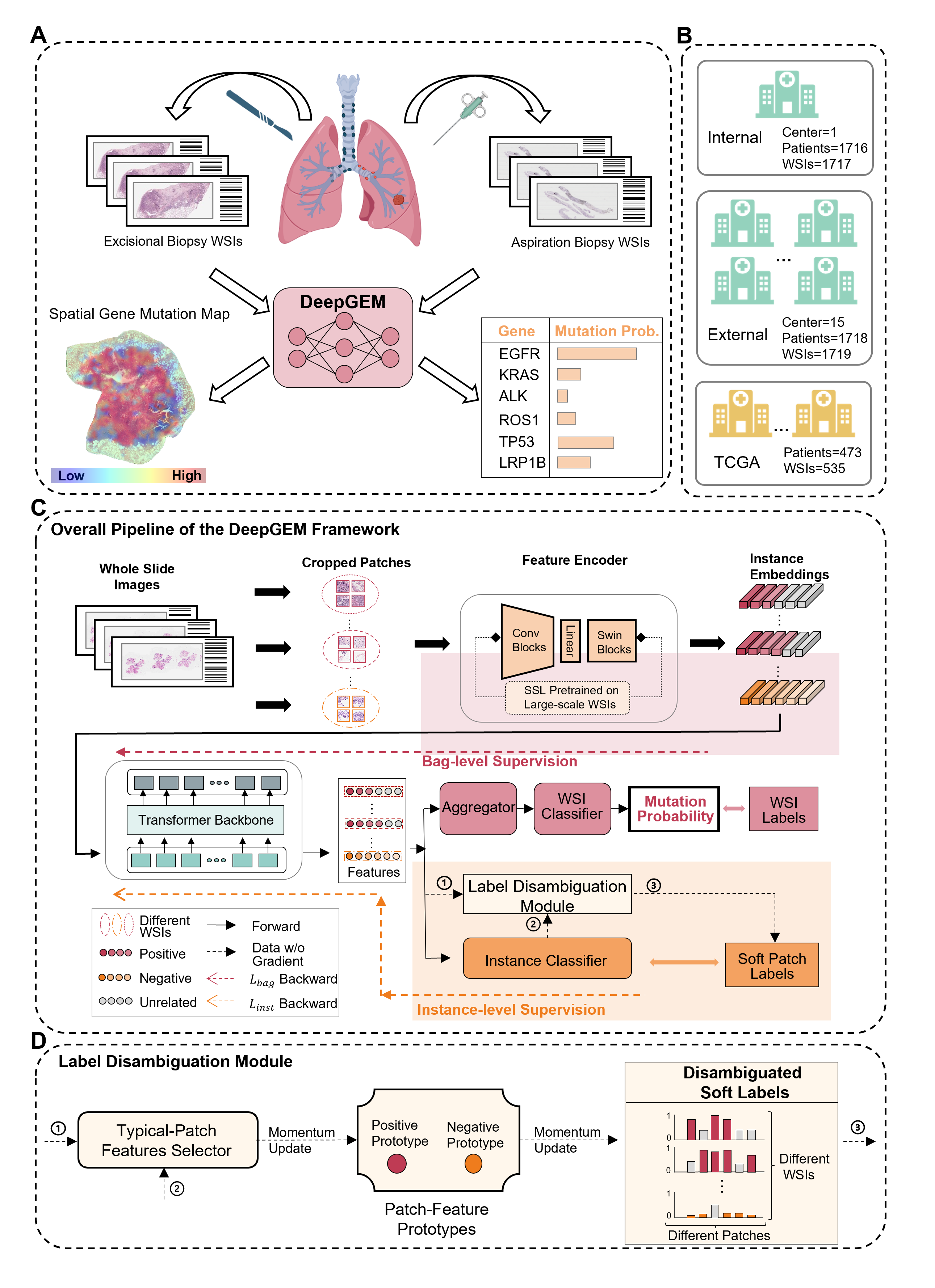
Deep multiple instance learning-enabled gene mutation prediction of lung cancer from histopathology images
We introduce a deep multiple instance learning-enabled artificial intelligence (AI) system (termed as DeepGEM for Deep GEne Mutation) to provide driver gene mutation prediction from routinely acquired histology slides without the need of manual annotation. Our model integrates a variety of advanced deep learning approaches and concepts, including self-supervised learning, weakly-supervised multiple instance learning, noisy label learning, transformer architecture, and label disambiguation. DeepGEM has shown the potential to be an economical, timely, and accurate gene mutation prediction tool for guiding clinical drug recommendations for lung cancer patients.
- DeepGEM
- System requirements
- Pathological Image preprocessing
- Install guide
- Dataset
- Disclaimer
- Coypright
- Contact
DeepGEM package requires only a standard computer with enough RAM and a NVIDIA GPU to support operations.
This tool is supported for Linux. The tool has been tested on the following systems:
- CentOS Linux release 8.2.2.2004
- Ubuntu 18.04.5 LTS
DeepGEM mainly depends on the Python scientific stack.
- The important packages including:
albumentations==1.1.0
efficientnet_pytorch==0.7.1
lifelines==0.26.4
numpy==1.21.6
opencv_contrib_python==4.8.0.74
opencv_python==4.4.0.46
opencv_python_headless==4.2.0.34
openslide_python==1.2.0
pandas==1.3.5
Pillow==8.4.0
rich==10.16.1
scikit_image==0.19.1
scikit_learn==1.0.1
torch==1.7.1+cu110
torchvision==0.8.2+cu110
tqdm==4.62.3
yacs==0.1.8
h5py==2.9.0
numba==0.44
torchtuples==0.2.0
- Install the modified timm library
./DeepGEM/requirements.txtdescribes more details of the requirements.
This code uses the centralized configs. Before using this code, a config file needs to be edited to assign necessary parameters. A sample config file named 'default.yaml' is provided as the reference.
./main/configs/default.yaml
1、First, in order to split the WSI into patches, execute the following script.
python ./data_prepare/step1_WSI_cropping.py \
--dataset ./sample_data/WSI \
--output ./sample_data/patch \
--scale 20 --patch_size 1120 --num_threads 16
2、Then, remove background patches.
python ./data_prepare/step2_get_patch_list_rmbk_mp.py \
--dataset ./sample_data/patch \
--output ./sample_data
3、Then, extract features from each patch. a pre-trained feature extractor can be utilized here (e.g. CTranspath trained in WSIs or EfficientNet-B0 trained on the ImageNet).
python ./data_prepare/step3_extract_feature.py --cfg ./configs/sample.yaml
3、Finally, combine features of one WSI.
python ./data_prepare/step4_merge_patch_feat.py --cfg configs/sample.yaml
docker pull skirophorion/deepgem:latest
The docker image file can be found at WeDrive
- Add docker image by loading the docker file:
docker load < deepgem.tar
docker run --name deepgem --gpus all -it --rm skirophorion/deepgem:latest /bin/bash
- If there are multiple GPUs in your device and you just want to use only one GPU:
docker run --name deepgem --gpus '"device=0"' -it --rm deepgem:latest /bin/bash
- Test directly because all files are contained in this docker image:
# Run commands in the folder `./DeepGEM`:
cd DeepGEM
# Sample Dataset
python ./main/test.py \
--cfg ./configs/sample.yaml \
--input_data ./sample_data/sample.csv \
--feat_dir ./sample_data/combined_feat \
--checkpoint ./checkpoints/DeepGEM_TCGA/modelTCGA_ExcisionalBiopsy_EGFR.pickle \
--gene EGFR \
--wsi_type ExcisionalBiopsy \
--save_testfile True \
# TCGA Dataset
python ./main/test.py \
--cfg ./configs/tcga.yaml \
--input_data ./data/tcga/tcga.pickle \
--feat_dir ./data/tcga/combined_feat \
--checkpoint ./checkpoints/DeepGEM_TCGA/modelTCGA_ExcisionalBiopsy_EGFR.pickle \
--gene EGFR \
--wsi_type ExcisionalBiopsy \
--save_testfile False \
# Internal Dataset
python ./main/test.py \
--cfg ./configs/internal.yaml \
--input_data ./data/internal/internal.pickle \
--feat_dir ./data/internal/combined_feat \
--checkpoint ./checkpoints/DeepGEM/model_ExcisionalBiopsy_EGFR.pickle \
--gene EGFR \
--wsi_type ExcisionalBiopsy \
--save_testfile False \
# External Dataset
python ./main/test.py \
--cfg ./configs/internal.yaml \
--input_data ./data/external/external.pickle \
--feat_dir ./data/external/combined_feat \
--checkpoint ./checkpoints/DeepGEM/model_ExcisionalBiopsy_EGFR.pickle \
--gene EGFR \
--wsi_type ExcisionalBiopsy \
--save_testfile False \
git clone https://github.com/TencentAILabHealthcare/DeepGEM.git
cd ./DeepGEM
conda create -n deepgem python=3.7
conda activate deepgem
pip install -r requirements.txt
pip install timm-0.5.4.tar
conda deactivate
- Run commands in the folder
./DeepGEM:
conda activate deepgem
# Sample Dataset
python ./main/test.py \
--cfg ./configs/sample.yaml \
--input_data ./sample_data/sample.csv \
--feat_dir ./sample_data/combined_feat \
--checkpoint ./checkpoints/DeepGEM_TCGA//modelTCGA_ExcisionalBiopsy_EGFR.pickle \
--gene EGFR \
--wsi_type ExcisionalBiopsy \
--save_testfile True \
3-Running (Needing to download the data first, please refer to the Dataset section for more details.):
- Run commands in the folder
./DeepGEM:
conda activate deepgem
# TCGA Dataset
python ./main/test.py \
--cfg ./configs/tcga.yaml \
--input_data ./data/tcga/tcga.pickle \
--feat_dir ./data/tcga/combined_feat \
--checkpoint ./checkpoints/DeepGEM_TCGA//modelTCGA_ExcisionalBiopsy_EGFR.pickle \
--gene EGFR \
--wsi_type ExcisionalBiopsy \
--save_testfile False \
# Internal Dataset
python ./main/test.py \
--cfg ./configs/internal.yaml \
--input_data ./data/internal/internal.pickle \
--feat_dir ./data/internal/combined_feat \
--checkpoint ./checkpoints/DeepGEM/model_ExcisionalBiopsy_EGFR.pickle \
--gene EGFR \
--wsi_type ExcisionalBiopsy \
--save_testfile False \
# External Dataset
python ./main/test.py \
--cfg ./configs/internal.yaml \
--input_data ./data/external/external.pickle \
--feat_dir ./data/external/combined_feat \
--checkpoint ./checkpoints/DeepGEM/model_ExcisionalBiopsy_EGFR.pickle \
--gene EGFR \
--wsi_type ExcisionalBiopsy \
--save_testfile False \
Example data are given in ./sample_data.
The DeepGEM source code, the training and test data, as well as the trained DeepGEM models, are also available on Zenodo, or WeDrive.
This tool is for research purpose and not approved for clinical use.
This tool is developed in Tencent AI Lab.
The copyright holder for this project is Tencent AI Lab.
All rights reserved.
Please get in touch with louisyuzhao@tencent.com if you have any questions.