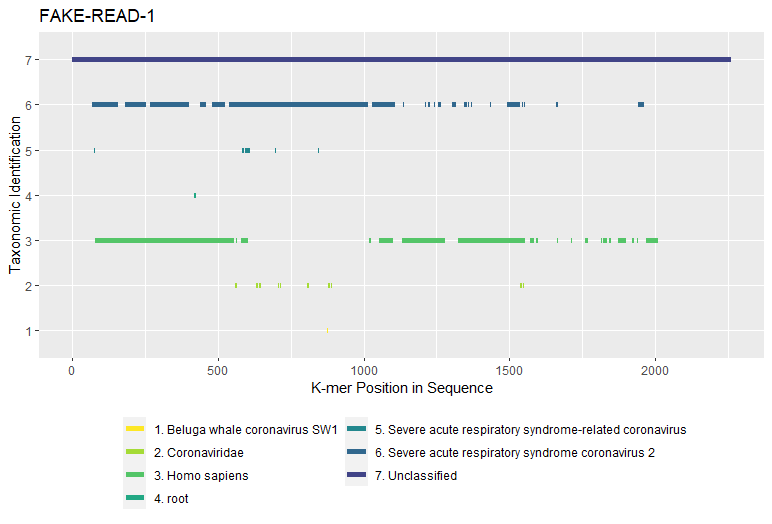
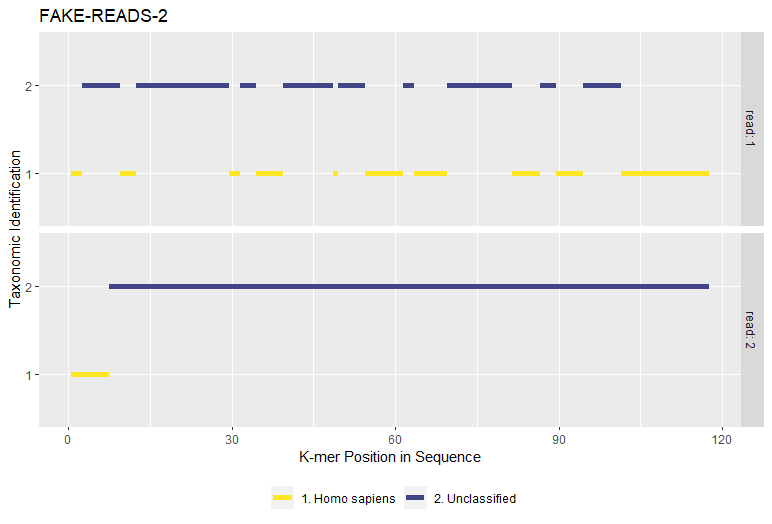
Create a conda environment and then run the following
sbatch -p NMLResearch -c 1 --mem=4G --wrap="wget -O - https://raw.githubusercontent.com/TheZetner/krakenreports/master/inst/exec/install.sh | bash"
- Sbatch is only necessary on slurm system
- Runs install.sh which does the following
- Installs...
- kraken2 from bioconda
- r-base from r
- r-base from r
- r-essentials from conda-forge
- r-xml from conda-forge
- Installs R Packages Remotes(CRAN) and Krakenreports(Github)
- Copies executable scripts to the bin/ folder of your Conda environment
- Installs...
To update krakenreports and its executable scripts run the following in your Conda environment.
sbatch -p NMLResearch -c 1 --mem=4G --wrap="wget -O - https://raw.githubusercontent.com/TheZetner/krakenreports/master/inst/exec/update.sh | bash"
runkraken.sh to run kraken2 via sbatch on a folder of fastq files
krakenreports.R to create plots and reports of the results
<PREFIX>_perseq.csv- Identified k-mer counts by taxonomy per read sequence
| Status | Sequence | TaxonomicName | Count |
|---|---|---|---|
| RUN1-kraken.tsv:C | FAKE-READ-1 | Unclassified | 1741 |
| RUN1-kraken.tsv:C | FAKE-READ-1 | Homo sapiens | 268 |
| RUN1-kraken.tsv:C | FAKE-READ-1 | Severe acute respiratory syndrome coronavirus 2 | 251 |
| RUN1-kraken.tsv:C | FAKE-READ-2 | Unclassified | 1219 |
| RUN1-kraken.tsv:C | FAKE-READ-2 | Homo sapiens | 246 |
| RUN1-kraken.tsv:C | FAKE-READ-2 | Severe acute respiratory syndrome coronavirus 2 | 187 |
| RUN1-kraken.tsv:C | FAKE-READ-2 | Coronaviridae | 5 |
| RUN1-kraken.tsv:C | FAKE-READ-2 | Severe acute respiratory syndrome-related coronavirus | 5 |
| RUN1-kraken.tsv:C | FAKE-READ-3 | Unclassified | 1068 |
| RUN1-kraken.tsv:C | FAKE-READ-3 | Homo sapiens | 352 |
| RUN1-kraken.tsv:C | FAKE-READ-3 | Severe acute respiratory syndrome coronavirus 2 | 305 |
| RUN1-kraken.tsv:C | FAKE-READ-3 | Severe acute respiratory syndrome-related coronavirus | 4 |
| RUN2-kraken.tsv:C | FAKE-READ-4 | Unclassified | 1108 |
| RUN2-kraken.tsv:C | FAKE-READ-4 | Homo sapiens | 338 |
| RUN2-kraken.tsv:C | FAKE-READ-4 | Severe acute respiratory syndrome coronavirus 2 | 241 |
<PREFIX>_allseq.csv- Identified k-mer counts by taxonomy for all
| TaxonomicName | Count |
|---|---|
| Unclassified | 11919 |
| Homo sapiens | 2850 |
| Severe acute respiratory syndrome coronavirus 2 | 2000 |
| Coronaviridae | 63 |
| Severe acute respiratory syndrome-related coronavirus | 36 |
| root | 5 |
| Beluga whale coronavirus SW1 | 2 |
Install alone via remotes:
Rscript --vanilla -e 'remotes::install_github("TheZetner/krakenreports")'
Sometimes it's worth linking files:
for i in `ls ../../<FOLDEROFFILES>/*-kraken.tsv`; do ln -s $i ./; done
Searching for human (9606) reads in kraken2 output:
grep -P "\s+9606\s+" -H *-kraken.tsv > human.tsv