This repo contains the official implementation for the paper Score-Based Generative Modeling through Stochastic Differential Equations
by Yang Song, Jascha Sohl-Dickstein, Diederik P. Kingma, Abhishek Kumar, Stefano Ermon, and Ben Poole
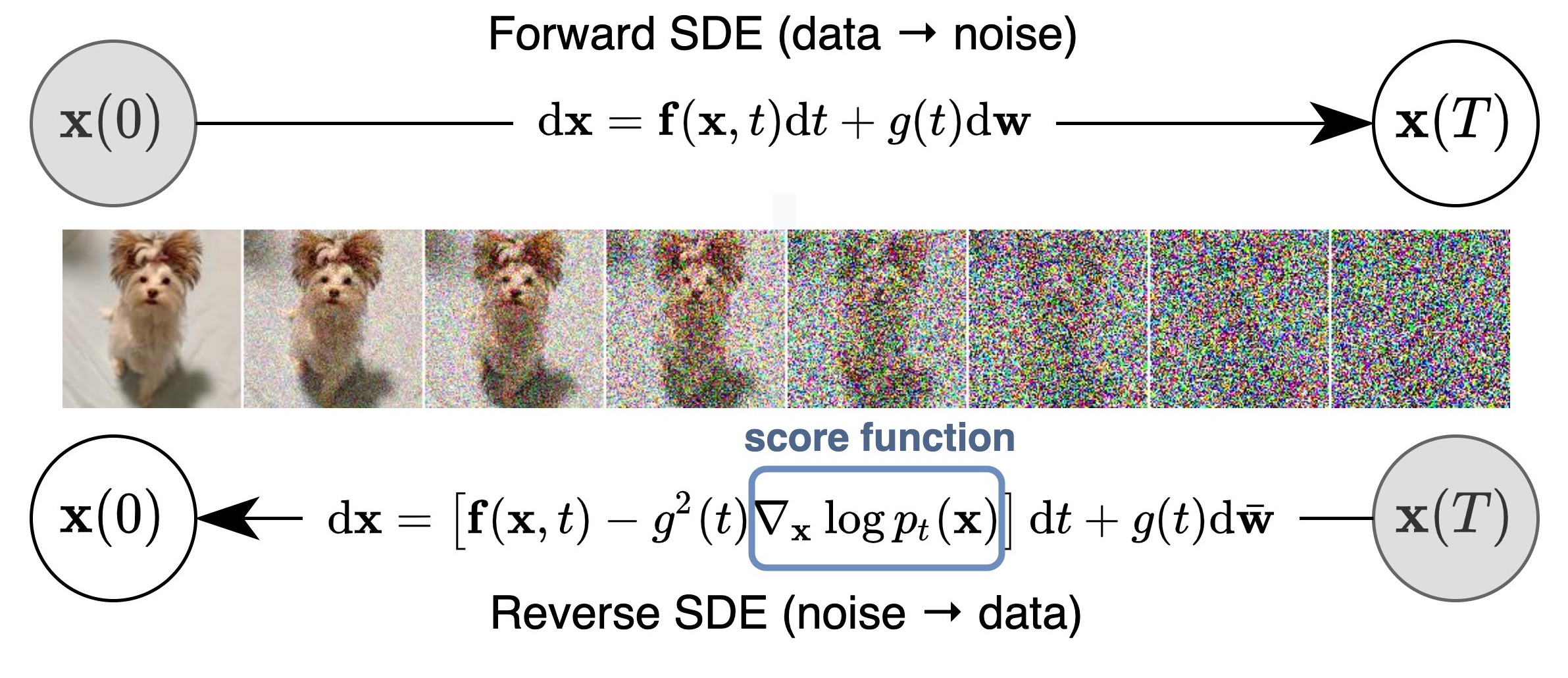
We propose a unified framework that generalizes and improves previous work on score-based generative models through the lens of stochastic differential equations (SDEs). In particular, we can transform data to a simple noise distribution with a continuous-time stochastic process described by an SDE. This SDE can be reversed for sample generation if we know the score of the marginal distributions at each intermediate time step, which can be estimated with score matching. The basic idea is captured in the figure below:
Our work enables a better understanding of existing approaches, new sampling algorithms, exact likelihood computation, uniquely identifiable encoding, latent code manipulation, and brings new conditional generation abilities to the family of score-based generative models.
All combined, we achieved an FID of 2.20 and an Inception score of 9.89 for unconditional generation on CIFAR-10, as well as high-fidelity generation of 1024px Celeba-HQ images. In addition, we obtained a likelihood value of 2.99 bits/dim on uniformly dequantized CIFAR-10 images.
Aside from the NCSN++ and DDPM++ models in our paper, this codebase also re-implements many previous score-based models all in one place, including NCSN from Generative Modeling by Estimating Gradients of the Data Distribution, NCSNv2 from Improved Techniques for Training Score-Based Generative Models, and DDPM from Denoising Diffusion Probabilistic Models.
It supports training new models, evaluating the sample quality and likelihoods of existing models. We carefully designed the code to be modular and easily extensible to new SDEs, predictors, or correctors.
Run the following to install a subset of necessary python packages for our code
pip install -r requirements.txtTrain and evaluate our models through main.py.
main.py:
--config: Training configuration.
(default: 'None')
--eval_folder: The folder name for storing evaluation results
(default: 'eval')
--mode: <train|eval>: Running mode: train or eval
--workdir: Working directory-
configis the path to the config file. Our prescribed config files are provided inconfigs/. They are formatted according toml_collectionsand should be quite self-explanatory. -
workdiris the path that stores all artifacts of one experiment, like checkpoints, samples, and evaluation results. -
eval_folderis the name of a subfolder inworkdirthat stores all artifacts of the evaluation process, like meta checkpoints for pre-emption prevention, image samples, and numpy dumps of quantitative results. -
modeis either "train" or "eval". When set to "train", it starts the training of a new model, or resumes the training of an old model if its meta-checkpoints (for resuming running after pre-emption in a cloud environment) exist inworkdir. When set to "eval", it can do an arbitrary combination of the following-
Evaluate the loss function on the test / validation dataset.
-
Generate a fixed number of samples and compute its Inception score, FID, or KID.
-
Compute the log-likelihood on the training or test dataset.
These functionalities can be configured through config files, or more conveniently, through the command-line support of the
ml_collectionspackage. For example, to generate samples and evaluate sample quality, supply the--config.eval.enable_samplingflag; to compute log-likelihoods, supply the--config.eval.enable_bpdflag, and specify--config.eval.dataset=train/testto indicate whether to compute the likelihoods on the training or test dataset. -
- New SDEs: inherent the
sde_lib.SDEabstract class and implement all abstract methods. Thediscretize()method is optional and the default is Euler-Maruyama discretization. Existing sampling methods and likelihood computation will automatically work for this new SDE. - New predictors: inherent the
sampling.Predictorabstract class, implement theupdate_fnabstract method, and register its name with@register_predictor. The new predictor can be directly used insampling.get_pc_samplerfor Predictor-Corrector sampling, and all other controllable generation methods incontrollable_generation.py. - New correctors: inherent the
sampling.Correctorabstract class, implement theupdate_fnabstract method, and register its name with@register_corrector. The new corrector can be directly used insampling.get_pc_sampler, and all other controllable generation methods incontrollable_generation.py.
Link: https://drive.google.com/drive/folders/10pQygNzF7hOOLwP3q8GiNxSnFRpArUxQ?usp=sharing
You may find two checkpoints for some models. The first checkpoint (with a smaller number) is the one that we reported FID scores in Table 3. The second checkpoint (with a larger number) is the one that we reported likelihood values and FIDs of black-box ODE samplers in Table 2. The former corresponds to the smallest FID during the course of training (every 50k iterations). The later is the last checkpoint during training.
If you find the code useful for your research, please consider citing
@inproceedings{
song2021scorebased,
title={Score-Based Generative Modeling through Stochastic Differential Equations},
author={Yang Song and Jascha Sohl-Dickstein and Diederik P Kingma and Abhishek Kumar and Stefano Ermon and Ben Poole},
booktitle={International Conference on Learning Representations},
year={2021},
url={https://openreview.net/forum?id=PxTIG12RRHS}
}This work is built upon some previous papers which might also interest you:
- Song, Yang, and Stefano Ermon. "Generative Modeling by Estimating Gradients of the Data Distribution." Proceedings of the 33rd Annual Conference on Neural Information Processing Systems. 2019.
- Song, Yang, and Stefano Ermon. "Improved techniques for training score-based generative models." Proceedings of the 34th Annual Conference on Neural Information Processing Systems. 2020.
- Ho, Jonathan, Ajay Jain, and Pieter Abbeel. "Denoising diffusion probabilistic models." Proceedings of the 34th Annual Conference on Neural Information Processing Systems. 2020.