Xuebin Qin, Hang Dai, Xiaobin Hu, Deng-Ping Fan*, Ling Shao, Luc Van Gool.
** (2022-Jul.-17)** Our paper, code and dataset are now officially released!!! Please check our project page for more details: Project Page.
** (2022-Jul.-5)** Our DIS work is now accepted by ECCV 2022, the code and dataset will be released before July 17th, 2022. Please be aware of our updates.
Download: Google Drive or Baidu Pan 提取码:rtgw
1.2 DIS dataset V2.0
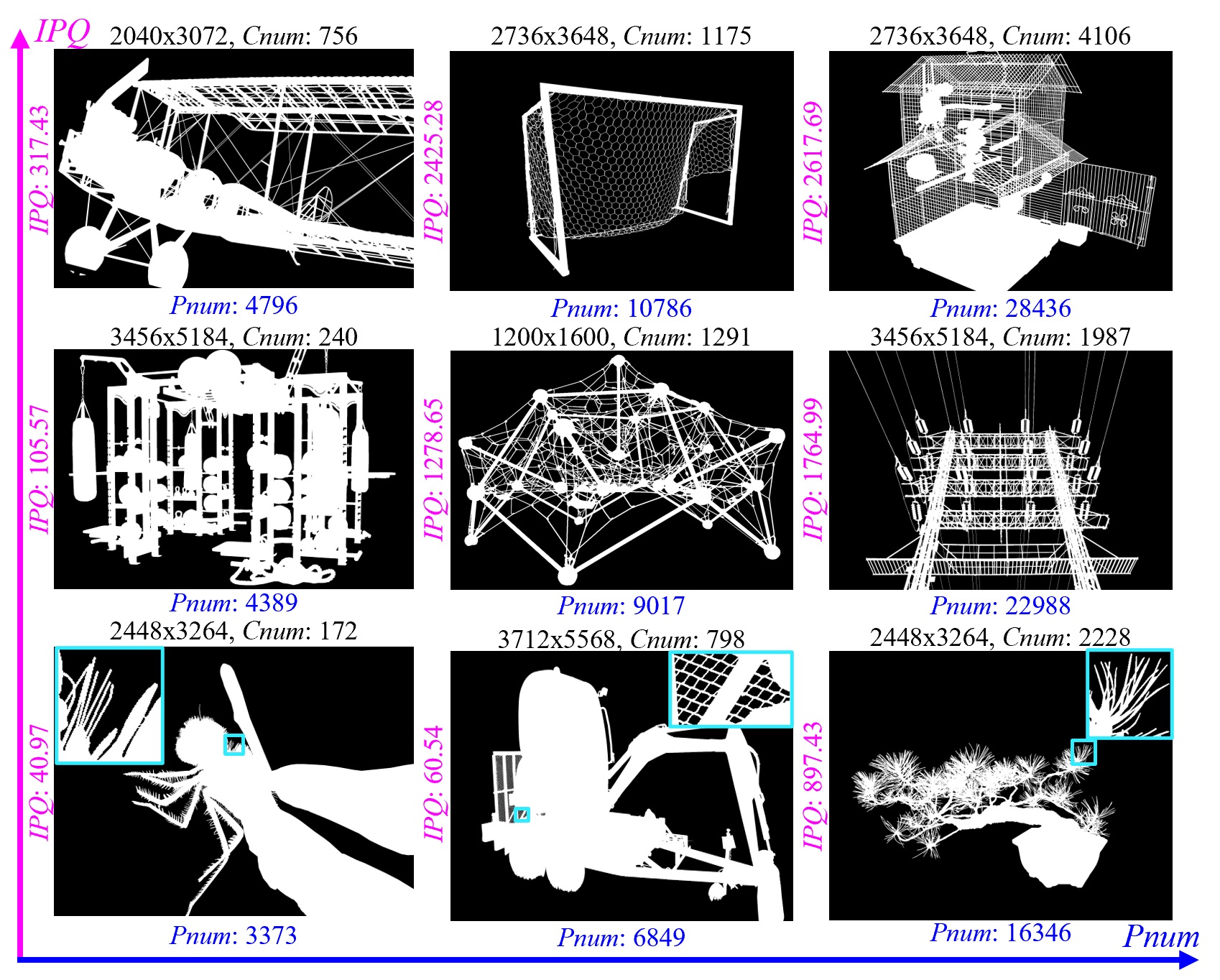
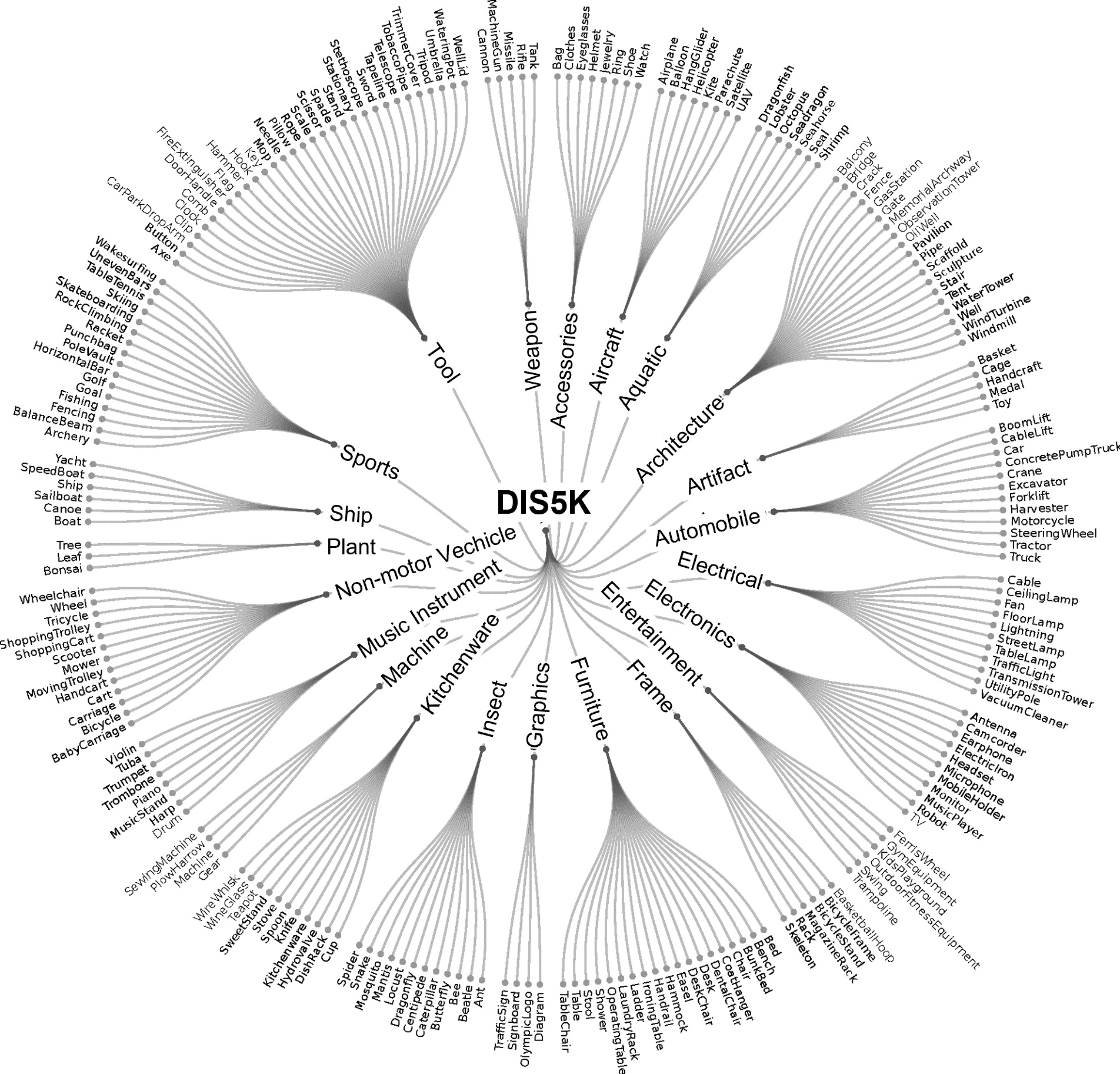
Although our DIS5K V1.0 includes samples from more than 200 categories, many categories, such as human, animals, cars and so on, in real world are not included. So the current version (v1.0) of our dataset may limit the robustness of the trained models. To build the comprehensive and large-scale highly accurate dichotomous image segmentation dataset, we are building our DIS dataset V2.0. The V2.0 will be released soon. Please stay tuned.
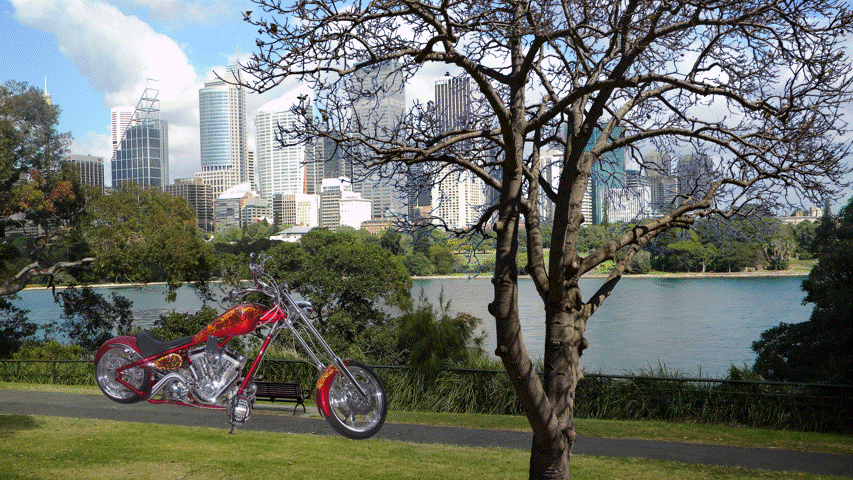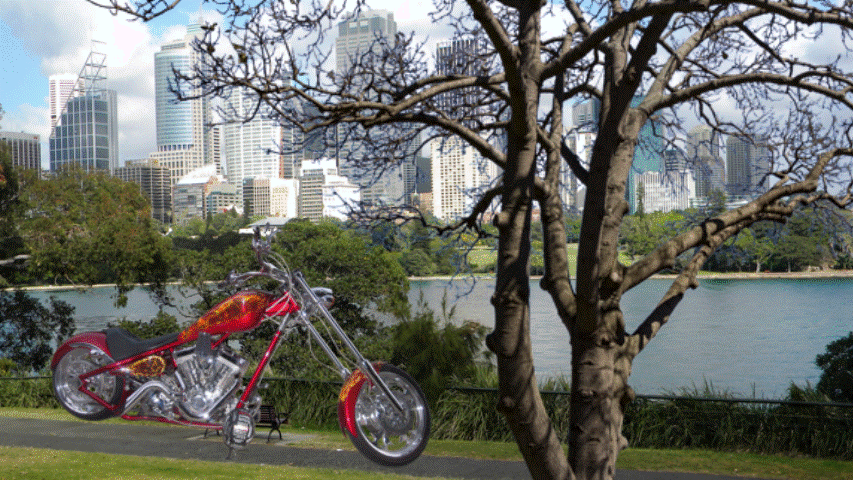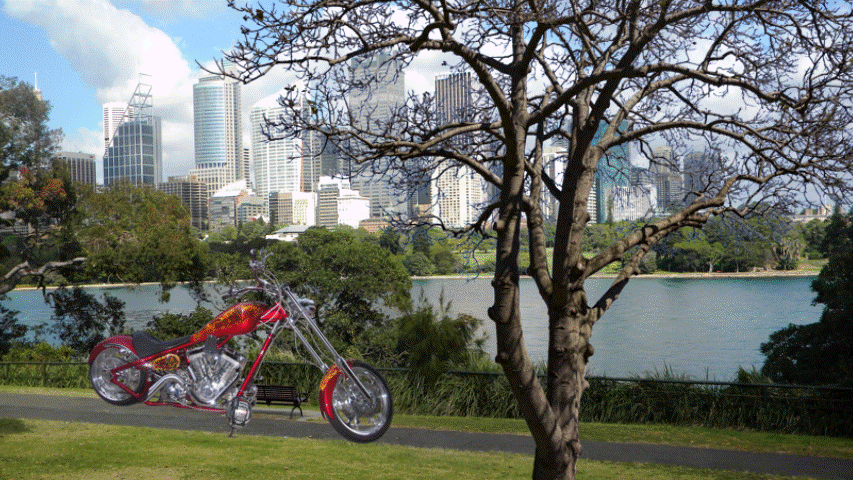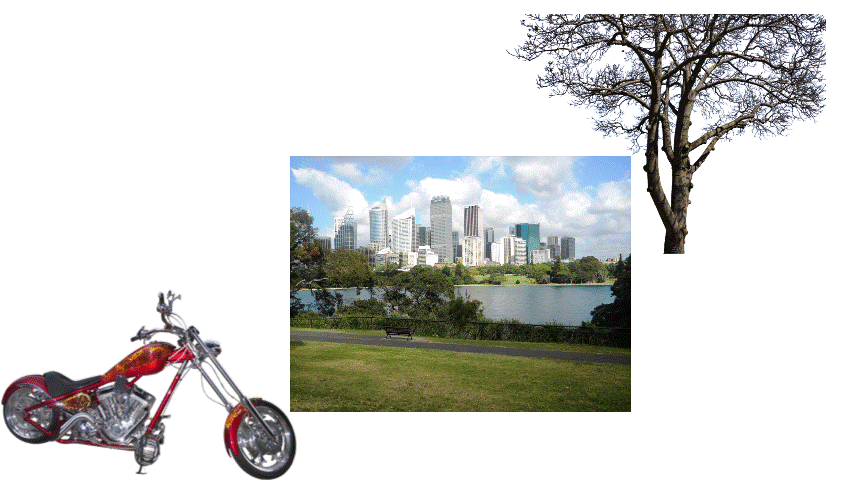
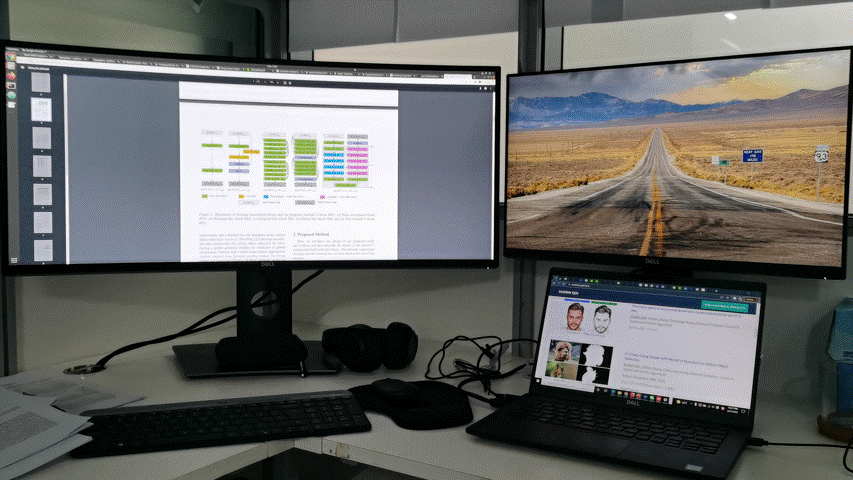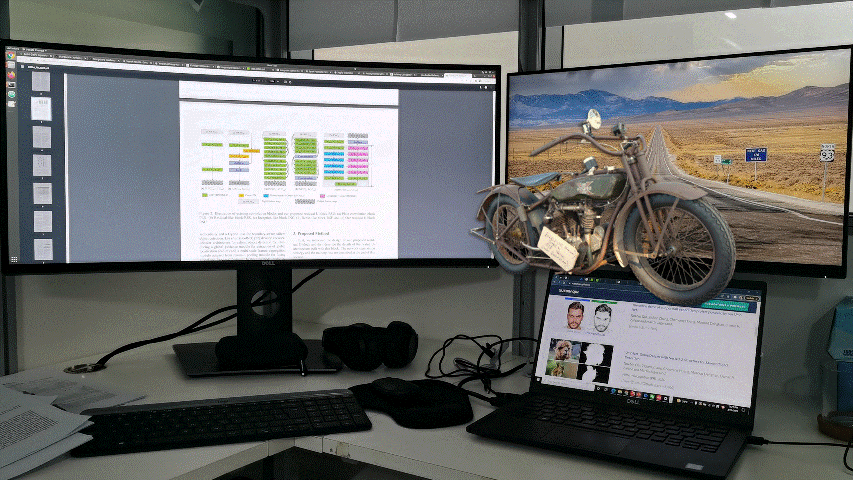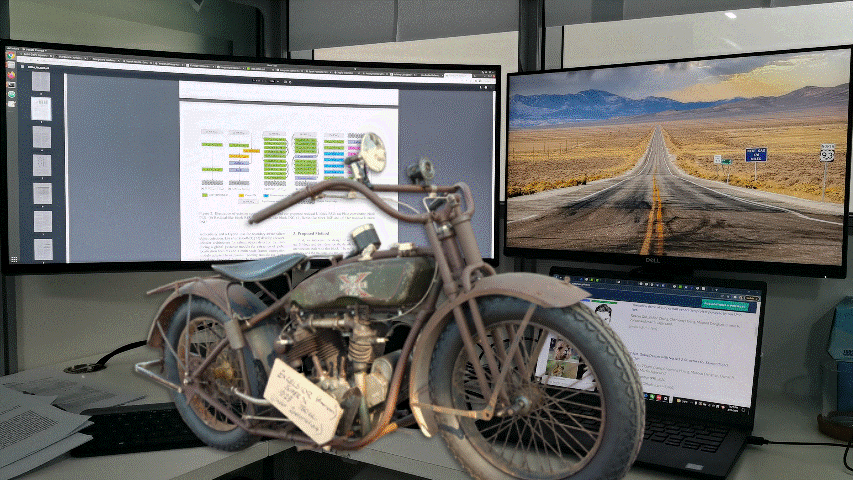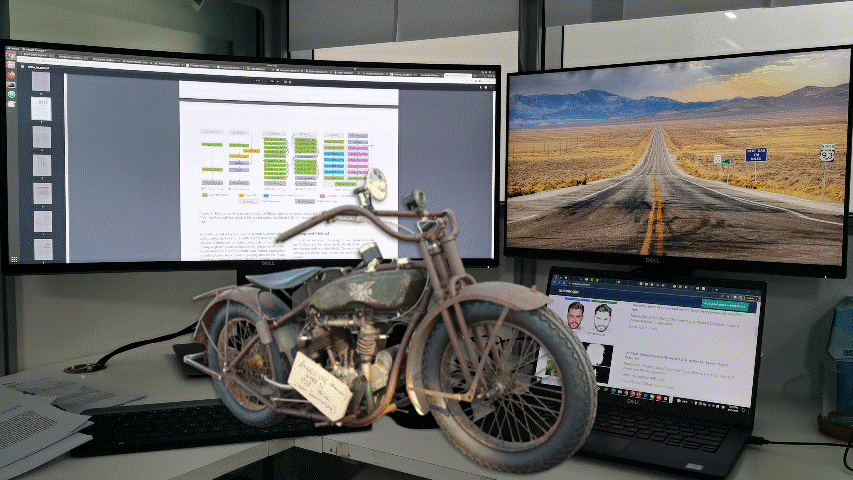
Samples from DIS dataset V2.0.

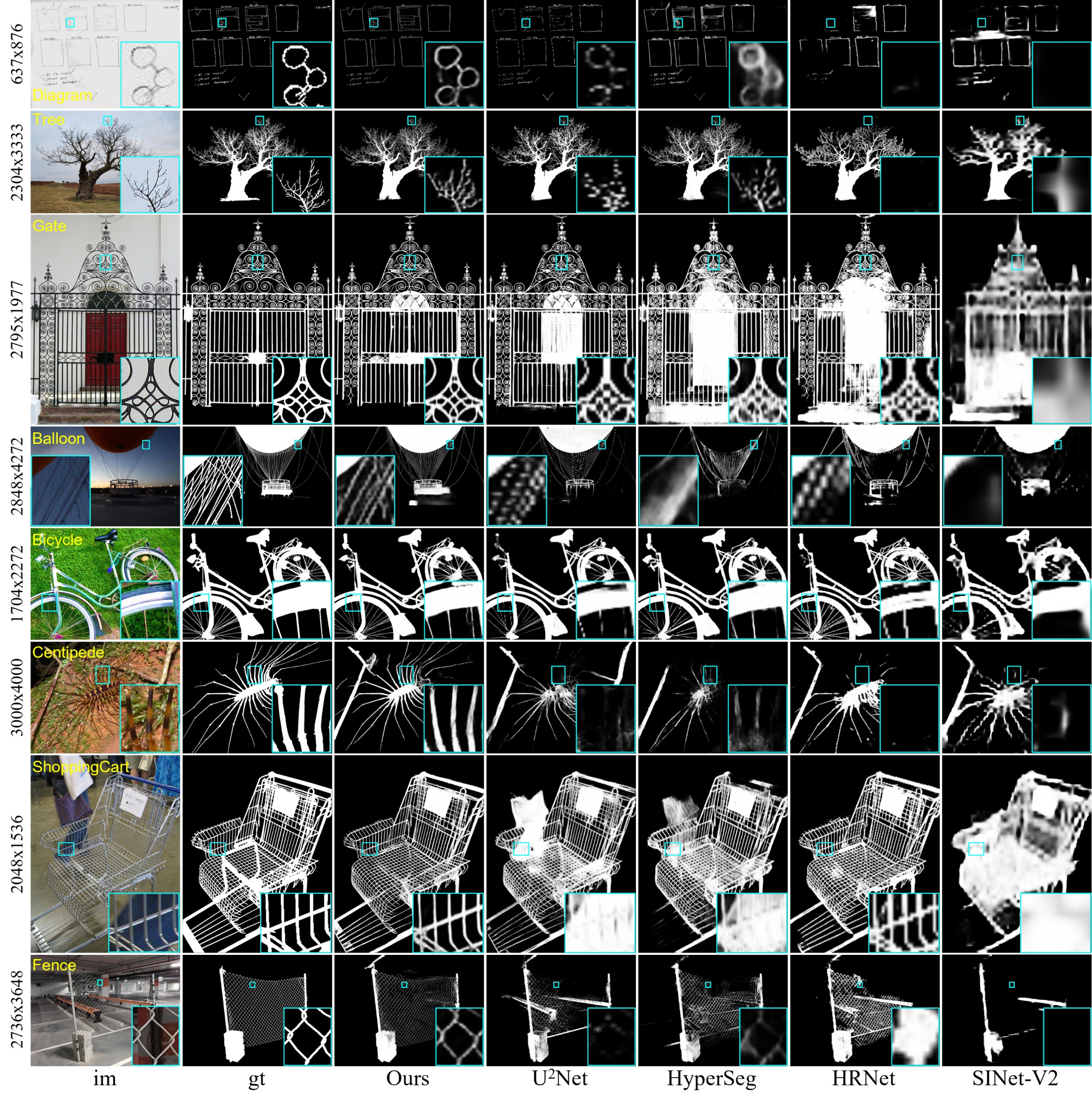
Predicted Maps, (Google Drive), (Baidu Pan 提取码:ph1d), of Our IS-Net and Other SOTAs
git clone https://github.com/xuebinqin/DIS.git
conda env create -f pytorch18.yml
Or you can check the requirements.txt to configure the dependancies.
(a) Open train_valid_inference_main.py, set the path of your to-be-inferenced train_datasets and valid_datasets, e.g., valid_datasets=[dataset_vd]
(b) Set the hypar["mode"] to "train"
(c) Create a new folder your_model_weights in the directory saved_models and set it as the hypar["model_path"] ="../saved_models/your_model_weights" and make sure hypar["valid_out_dir"](line 668) is set to "", otherwise the prediction maps of the validation stage will be saved to that directory, which will slow the training speed down
(d) Run
python train_valid_inference_main.py
(a). Download the pre-trained weights (for fair academic comparisons only, the optimized model for engineering or common use will be released soon) isnet.pth from (Google Drive) or (Baidu Pan 提取码:xbfk) and store isnet.pth in saved_models/IS-Net
(b) Open train_valid_inference_main.py, set the path of your to-be-inferenced valid_datasets, e.g., valid_datasets=[dataset_te1, dataset_te2, dataset_te3, dataset_te4]
(c) Set the hypar["mode"] to "valid"
(d) Set the output directory of your predicted maps, e.g., hypar["valid_out_dir"] = "../DIS5K-Results-test"
(e) Run
python train_valid_inference_main.py
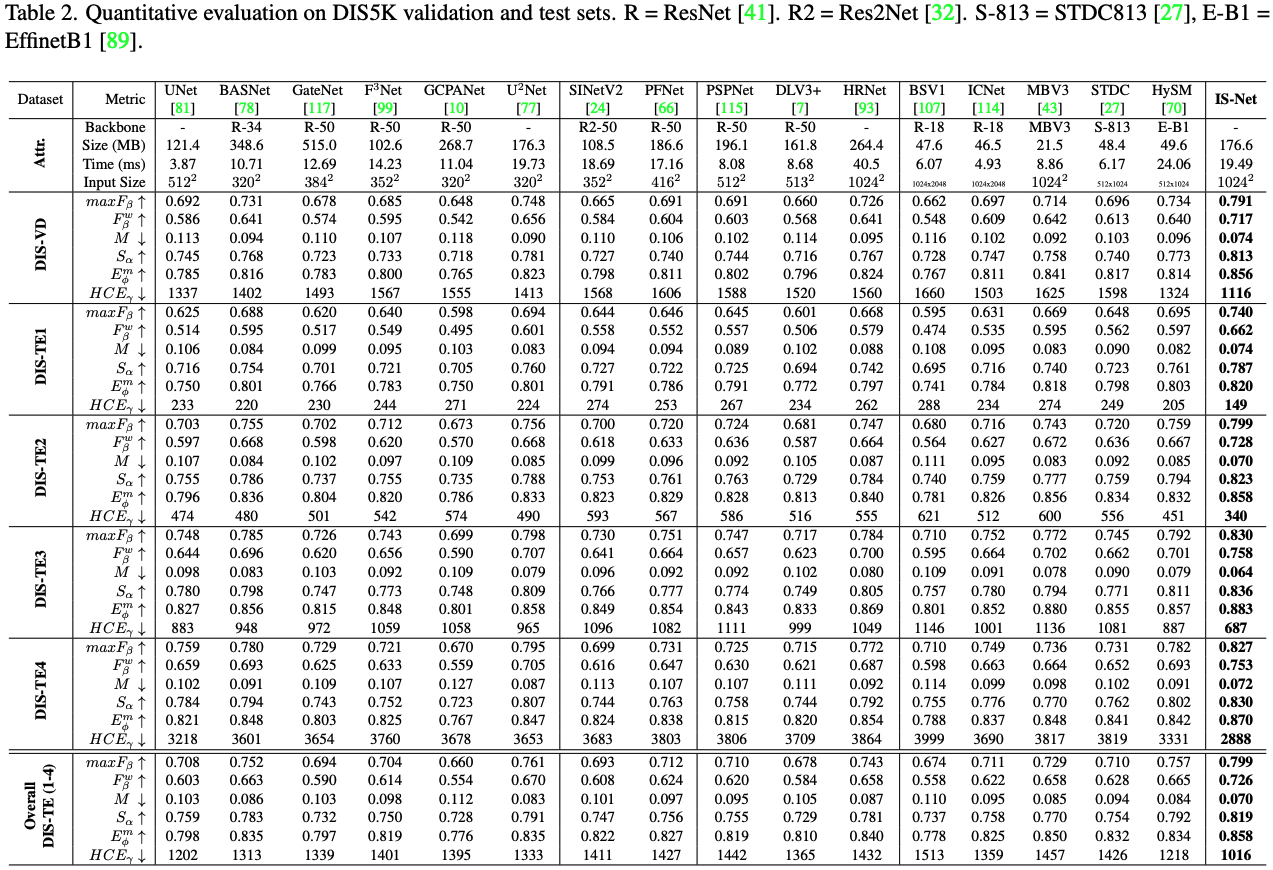
Set the ground truth directory gt_root and the prediction directory pred_root. To reduce the time costs for computing HCE, the skeletion of the DIS5K dataset can be pre-computed and stored in gt_ske_root. If gt_ske_root="", the HCE code will compute the skeleton online which usually takes a lot for time for large size ground truth. Then, run python hce_metric_main.py. Other metrics are evaluated based on the SOCToolbox.
Our code and evaluation metric use Apache License 2.0. The Terms of use for our DIS5K dataset is provided as DIS5K-Dataset-Terms-of-Use.pdf.
We would like to thank Dr. Ibrahim Almakky for his helps in implementing the dataloader cache machanism of loading large-size training samples and Jiayi Zhu for his efforts in re-organizing our code and dataset.
@InProceedings{qin2022,
author={Xuebin Qin and Hang Dai and Xiaobin Hu and Deng-Ping Fan and Ling Shao and Luc Van Gool},
title={Highly Accurate Dichotomous Image Segmentation},
booktitle={ECCV},
year={2022}
}
@InProceedings{Qin_2020_PR,
title = {U2-Net: Going Deeper with Nested U-Structure for Salient Object Detection},
author = {Qin, Xuebin and Zhang, Zichen and Huang, Chenyang and Dehghan, Masood and Zaiane, Osmar and Jagersand, Martin},
journal = {Pattern Recognition},
volume = {106},
pages = {107404},
year = {2020}
}
@InProceedings{Qin_2019_CVPR,
author = {Qin, Xuebin and Zhang, Zichen and Huang, Chenyang and Gao, Chao and Dehghan, Masood and Jagersand, Martin},
title = {BASNet: Boundary-Aware Salient Object Detection},
booktitle = {The IEEE Conference on Computer Vision and Pattern Recognition (CVPR)},
month = {June},
year = {2019}
}
@article{qin2021boundary,
title={Boundary-aware segmentation network for mobile and web applications},
author={Qin, Xuebin and Fan, Deng-Ping and Huang, Chenyang and Diagne, Cyril and Zhang, Zichen and Sant'Anna, Adri{\`a} Cabeza and Suarez, Albert and Jagersand, Martin and Shao, Ling},
journal={arXiv preprint arXiv:2101.04704},
year={2021}
}