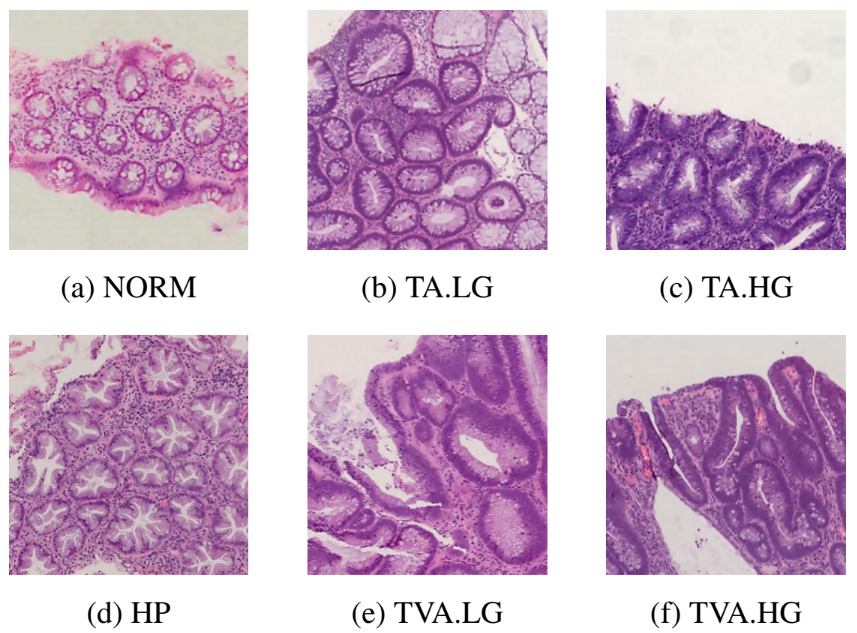
UniToPatho is an annotated dataset of 9536 hematoxylin and eosin stained patches extracted from 292 whole-slide images, meant for training deep neural networks for colorectal polyps classification and adenomas grading. The slides are acquired through a Hamamatsu Nanozoomer S210 scanner at 20× magnification (0.4415 μm/px). Each slide belongs to a different patient and is annotated by expert pathologists, according to six classes as follows:
- NORM - Normal tissue;
- HP - Hyperplastic Polyp;
- TA.HG - Tubular Adenoma, High-Grade dysplasia;
- TA.LG - Tubular Adenoma, Low-Grade dysplasia;
- TVA.HG - Tubulo-Villous Adenoma, High-Grade dysplasia;
- TVA.LG - Tubulo-Villous Adenoma, Low-Grade dysplasia.
You can download UniToPatho from IEEE-DataPort
We provide a PyTorch compatible dataset class and ECVL compatible dataloader. For example usage see Example.ipynb
If you use this dataset, please make sure to cite the related work:
@article{barbano2021unitopatho,
title={UniToPatho, a labeled histopathological dataset for colorectal polyps classification and adenoma dysplasia grading},
author={Barbano, Carlo Alberto and Perlo, Daniele and Tartaglione, Enzo and Fiandrotti, Attilio and Bertero, Luca and Cassoni, Paola and Grangetto, Marco},
journal={arXiv preprint arXiv:2101.09991},
year={2021}
}
and the dataset entry
@data{9fsv-tm25-21,
doi = {10.21227/9fsv-tm25},
url = {https://dx.doi.org/10.21227/9fsv-tm25},
author = {Luca Bertero; Carlo Alberto Barbano; Daniele Perlo; Enzo Tartaglione; Paola Cassoni; Marco Grangetto; Attilio Fiandrotti; Alessandro Gambella; Luca Cavallo },
publisher = {IEEE Dataport},
title = {UNITOPATHO},
year = {2021}
}