| fgivenx: | Functional Posterior Plotter |
|---|---|
| Author: | Will Handley |
| Version: | 2.2.0 |
| Homepage: | https://github.com/williamjameshandley/fgivenx |
| Documentation: | http://fgivenx.readthedocs.io/ |
fgivenx is a python package for plotting posteriors of functions. It is
currently used in astronomy, but will be of use to any scientists performing
Bayesian analyses which have predictive posteriors that are functions.
This package allows one to plot a predictive posterior of a function,
dependent on sampled parameters. We assume one has a Bayesian posterior
Post(theta|D,M) described by a set of posterior samples {theta_i}~Post.
If there is a function parameterised by theta y=f(x;theta), then this script
will produce a contour plot of the conditional posterior P(y|x,D,M) in the
(x,y) plane.
The driving routines are fgivenx.plot_contours, fgivenx.plot_lines and
fgivenx.plot_dkl. The code is compatible with getdist, and has a loading function
provided by fgivenx.samples_from_getdist_chains.
Users can install using pip:
pip install fgivenxfrom source:
git clone https://github.com/williamjameshandley/fgivenx
cd fgivenx
python setup.py install --useror for those on Arch linux it is available on the AUR
You can check that things are working by running the test suite (You may
encounter warnings if the optional dependency joblib is not installed):
pip install pytest pytest-runner pytest-mpl
export MPLBACKEND=Agg
pytest <fgivenx-install-location>
# or, equivalently
git clone https://github.com/williamjameshandley/fgivenx
cd fgivenx
python setup.py testCheck the dependencies listed in the next section are installed. You can then use the
fgivenx module from your scripts.
Some users of OSX or Anaconda may find QueueManagerThread errors if Pillow is not installed (run pip install pillow).
If you want to use parallelisation, have progress bars or getdist compatibility you should install the additional optional dependencies:
pip install joblib tqdm getdist
# or, equivalently
pip install -r requirements.txtYou may encounter warnings if you don't have the optional dependency joblib
installed.
Basic requirements:
- Python 2.7+ or 3.4+
- matplotlib
- numpy
- scipy
Documentation:
Tests:
Optional extras:
- joblib (parallelisation) [+ pillow on some systems]
- tqdm (progress bars)
- getdist (reading of getdist compatible files)
Full Documentation is hosted at ReadTheDocs. To build your own local copy of the documentation you'll need to install sphinx. You can then run:
cd docs
make htmlIf you use fgivenx to generate plots for a publication, please cite
as:
Handley, (2018). fgivenx: A Python package for functional posterior plotting . Journal of Open Source Software, 3(28), 849, https://doi.org/10.21105/joss.00849
or using the BibTeX:
@article{fgivenx,
doi = {10.21105/joss.00849},
url = {http://dx.doi.org/10.21105/joss.00849},
year = {2018},
month = {Aug},
publisher = {The Open Journal},
volume = {3},
number = {28},
author = {Will Handley},
title = {fgivenx: Functional Posterior Plotter},
journal = {The Journal of Open Source Software}
}import numpy
import matplotlib.pyplot as plt
from fgivenx import plot_contours, plot_lines, plot_dkl
# Model definitions
# =================
# Define a simple straight line function, parameters theta=(m,c)
def f(x, theta):
m, c = theta
return m * x + c
numpy.random.seed(1)
# Posterior samples
nsamples = 1000
ms = numpy.random.normal(loc=-5, scale=1, size=nsamples)
cs = numpy.random.normal(loc=2, scale=1, size=nsamples)
samples = numpy.array([(m, c) for m, c in zip(ms, cs)]).copy()
# Prior samples
ms = numpy.random.normal(loc=0, scale=5, size=nsamples)
cs = numpy.random.normal(loc=0, scale=5, size=nsamples)
prior_samples = numpy.array([(m, c) for m, c in zip(ms, cs)]).copy()
# Set the x range to plot on
xmin, xmax = -2, 2
nx = 100
x = numpy.linspace(xmin, xmax, nx)
# Set the cache
cache = 'cache/test'
prior_cache = cache + '_prior'
# Plotting
# ========
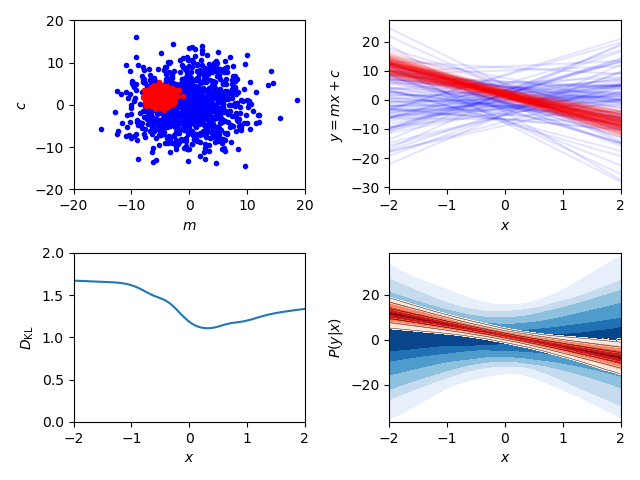
fig, axes = plt.subplots(2, 2)
# Sample plot
# -----------
ax_samples = axes[0, 0]
ax_samples.set_ylabel(r'$c$')
ax_samples.set_xlabel(r'$m$')
ax_samples.plot(prior_samples.T[0], prior_samples.T[1], 'b.')
ax_samples.plot(samples.T[0], samples.T[1], 'r.')
# Line plot
# ---------
ax_lines = axes[0, 1]
ax_lines.set_ylabel(r'$y = m x + c$')
ax_lines.set_xlabel(r'$x$')
plot_lines(f, x, prior_samples, ax_lines, color='b', cache=prior_cache)
plot_lines(f, x, samples, ax_lines, color='r', cache=cache)
# Predictive posterior plot
# -------------------------
ax_fgivenx = axes[1, 1]
ax_fgivenx.set_ylabel(r'$P(y|x)$')
ax_fgivenx.set_xlabel(r'$x$')
cbar = plot_contours(f, x, prior_samples, ax_fgivenx,
colors=plt.cm.Blues_r, lines=False,
cache=prior_cache)
cbar = plot_contours(f, x, samples, ax_fgivenx, cache=cache)
# DKL plot
# --------
ax_dkl = axes[1, 0]
ax_dkl.set_ylabel(r'$D_\mathrm{KL}$')
ax_dkl.set_xlabel(r'$x$')
ax_dkl.set_ylim(bottom=0, top=2.0)
plot_dkl(f, x, samples, prior_samples, ax_dkl,
cache=cache, prior_cache=prior_cache)
ax_lines.get_shared_x_axes().join(ax_lines, ax_fgivenx, ax_samples)
fig.tight_layout()
fig.savefig('plot.png')import numpy
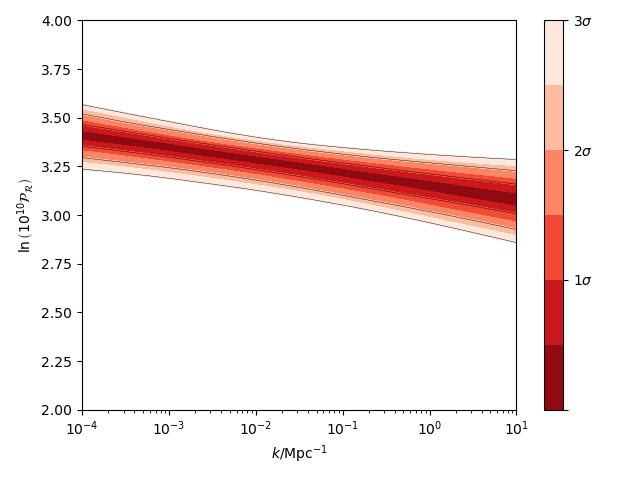
import matplotlib.pyplot as plt
from fgivenx import plot_contours, samples_from_getdist_chains
file_root = './plik_HM_TT_lowl/base_plikHM_TT_lowl'
samples, weights = samples_from_getdist_chains(['logA', 'ns'], file_root)
def PPS(k, theta):
logA, ns = theta
return logA + (ns - 1) * numpy.log(k)
k = numpy.logspace(-4,1,100)
cbar = plot_contours(PPS, k, samples, weights=weights)
cbar = plt.colorbar(cbar,ticks=[0,1,2,3])
cbar.set_ticklabels(['',r'$1\sigma$',r'$2\sigma$',r'$3\sigma$'])
plt.xscale('log')
plt.ylim(2,4)
plt.ylabel(r'$\ln\left(10^{10}\mathcal{P}_\mathcal{R}\right)$')
plt.xlabel(r'$k / {\rm Mpc}^{-1}$')
plt.tight_layout()
plt.savefig('planck.png')Want to contribute to fgivenx? Awesome!
There are many ways you can contribute via the
[GitHub repository](https://github.com/williamjameshandley/fgivenx),
see below.
Open an issue to report bugs or to propose new features.
Pull requests are very welcome. Note that if you are going to propose drastic changes, be sure to open an issue for discussion first, to make sure that your PR will be accepted before you spend effort coding it.
| v2.2.0: | Paper accepted |
|---|---|
| v2.1.17: | 100% coverage |
| v2.1.16: | Tests fixes |
| v2.1.15: | Additional plot tests |
| v2.1.13: | Further bug fix in test suite for image comparison |
| v2.1.12: | Bug fix in test suite for image comparison |
| v2.1.11: | Documentation upgrades |
| v2.1.10: | Added changelog |