This repo contains pre-trained Meta-matching models. If you want to train your own meta-matching model from scratch, please visit our CBIG repo.
- He, T., An, L., Chen, P., Chen, J., Feng, J., Bzdok, D., Holmes, A.J., Eickhoff, S.B. and Yeo, B.T., 2022. Meta-matching as a simple framework to translate phenotypic predictive models from big to small data, Nature Neuroscience 25, 795-804.
- Chen, P., An, L., Wulan, N., Zhang, C., Zhang, S., Ooi, L. Q. R., ... & Yeo, B. T. (2024). Multilayer meta-matching: translating phenotypic prediction models from multiple datasets to small data. Imaging Neuroscience, 2, 1-22.
- Wulan, N., An, L., Zhang, C., Kong, R., Chen, P., Bzdok, D., ... & Yeo, B. T. (2024). Translating phenotypic prediction models from big to small anatomical MRI data using meta-matching. bioRxiv, 2023-12.
There is significant interest in using brain imaging to predict phenotypes, such as cognitive performance or clinical outcomes. However, most prediction studies are underpowered. We propose a simple framework - meta-matching - to translate predictive models from large-scale datasets to new unseen non-brain-imaging phenotypes in small-scale studies. The key consideration is that a unique phenotype from a boutique study likely correlates with (but is not the same as) related phenotypes in some large-scale dataset. Meta-matching exploits these correlations to boost prediction in the boutique study.
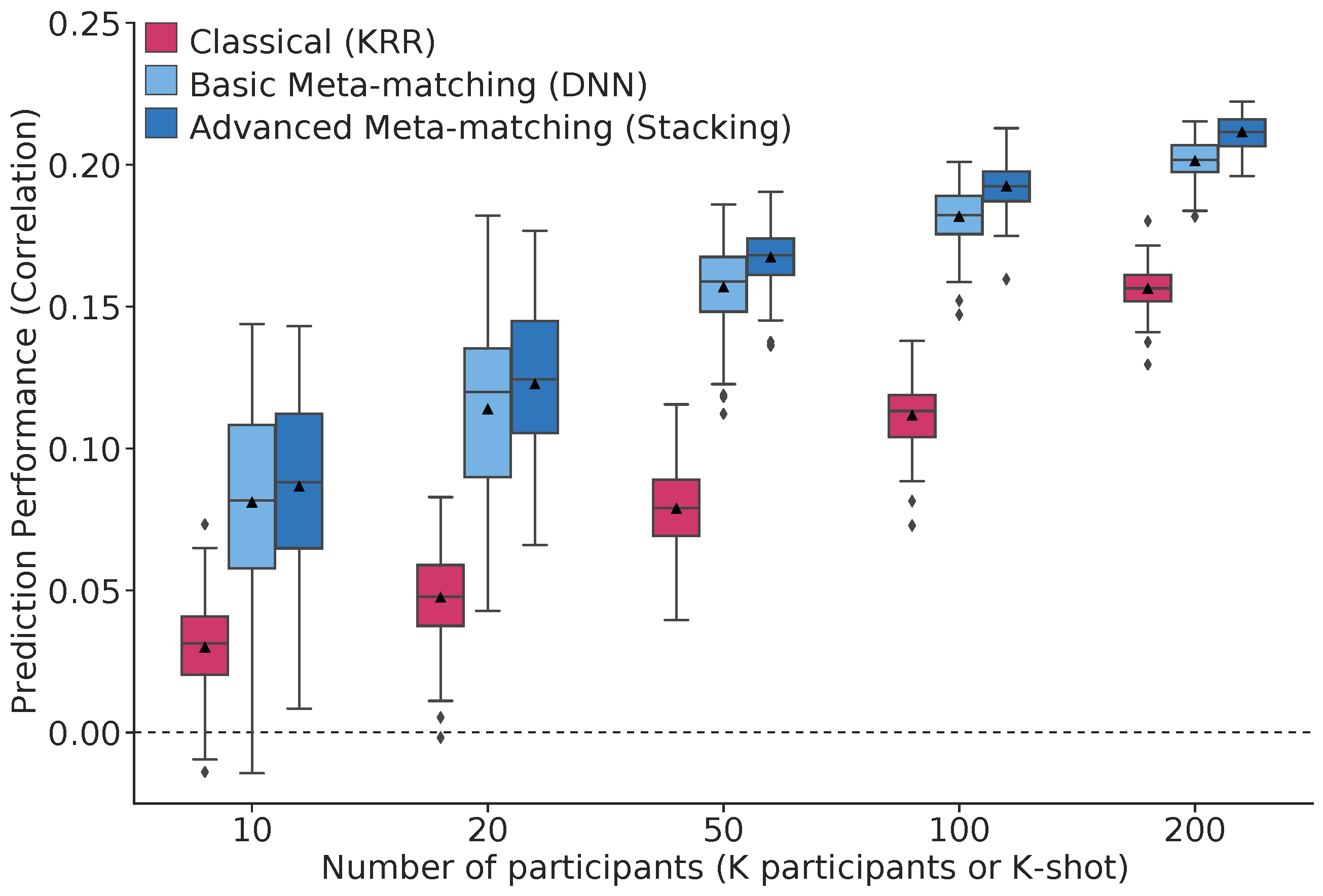
For example, we applied meta-matching to predict non-brain-imaging phenotypes from resting-state functional connectivity. Using the UK Biobank (N=36,848) and HCP (N=1,019) datasets, we demonstrate that meta-matching can greatly boost the prediction of new phenotypes in small independent datasets in many scenarios. For example, translating a UK Biobank model to 100 HCP participants yields an 8-fold improvement in variance explained with an average absolute gain of 4.0% (min=-0.2%, max=16.0%) across 35 phenotypes.
We have released multi-modality meta-matching models for both rs-fMRI and T1 data, check usage if you are interested.
Please check the detailed readme under each folder.
rs-fMRI folder contains meta-matching models for resting-state functional MRI (rs-fMRI) data
T1 folder contains meta-matching models for T1-weighted image data
See our LICENSE file for license rights and limitations (MIT).
Please contact He Tong at hetong1115@gmail.com, Lijun An at anlijun.cn@gmail.com, Pansheng Chen at chenpansheng@gmail.com and Thomas Yeo at yeoyeo02@gmail.com.
Happy researching!